Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
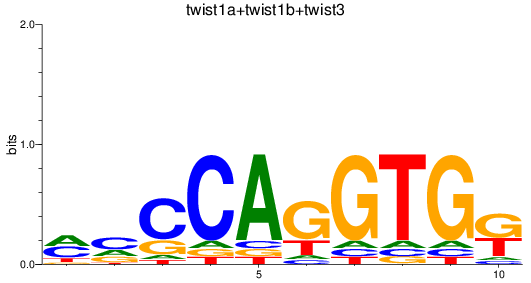
Results for twist1a+twist1b+twist3
Z-value: 0.36
Transcription factors associated with twist1a+twist1b+twist3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
twist3
|
ENSDARG00000019646 | twist3 |
|
twist1a
|
ENSDARG00000030402 | twist family bHLH transcription factor 1a |
|
twist1b
|
ENSDARG00000076010 | twist family bHLH transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| twist1b | dr11_v1_chr16_+_19732543_19732543 | -0.50 | 2.8e-02 | Click! |
| twist1a | dr11_v1_chr19_-_2231146_2231188 | 0.44 | 5.8e-02 | Click! |
| twist3 | dr11_v1_chr23_+_2669_2669 | -0.40 | 9.1e-02 | Click! |
Activity profile of twist1a+twist1b+twist3 motif
Sorted Z-values of twist1a+twist1b+twist3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_46216703 | 0.40 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr21_-_43666420 | 0.37 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr14_-_31087830 | 0.35 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr12_-_11294979 | 0.35 |
ENSDART00000148850
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr5_-_32505109 | 0.35 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr20_+_36623807 | 0.33 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr17_+_1360192 | 0.33 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr23_-_21471022 | 0.33 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr13_-_12602920 | 0.31 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr20_-_2134620 | 0.31 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr20_-_22193190 | 0.30 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr6_-_55423220 | 0.29 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr13_-_31296358 | 0.27 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr22_-_5752009 | 0.24 |
ENSDART00000190052
|
bcdin3d
|
BCDIN3 domain containing |
| chr5_+_53482597 | 0.24 |
ENSDART00000180333
|
BX323994.1
|
|
| chr13_+_25486608 | 0.23 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr22_-_26251563 | 0.23 |
ENSDART00000060888
ENSDART00000142821 |
ccdc130
|
coiled-coil domain containing 130 |
| chr11_-_34232906 | 0.23 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr6_-_10728921 | 0.22 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr13_-_9119867 | 0.22 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr4_+_77973876 | 0.22 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr6_-_10728057 | 0.20 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr5_-_11809404 | 0.20 |
ENSDART00000132564
|
nf2a
|
neurofibromin 2a (merlin) |
| chr18_-_35842554 | 0.19 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr2_-_10896745 | 0.19 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr13_-_36579086 | 0.18 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr15_-_1590858 | 0.17 |
ENSDART00000081875
|
nnr
|
nanor |
| chr17_-_40876680 | 0.15 |
ENSDART00000127200
|
supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr23_-_36313431 | 0.15 |
ENSDART00000125860
|
nfe2
|
nuclear factor, erythroid 2 |
| chr7_+_4671483 | 0.15 |
ENSDART00000111390
|
si:ch211-225k7.6
|
si:ch211-225k7.6 |
| chr24_+_39660124 | 0.14 |
ENSDART00000066500
|
stub1
|
STIP1 homology and U-Box containing protein 1 |
| chr3_+_58472305 | 0.14 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr4_+_33830944 | 0.13 |
ENSDART00000170238
|
znf1026
|
zinc finger protein 1026 |
| chr8_+_247163 | 0.13 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr23_+_2914577 | 0.13 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr10_+_40737540 | 0.11 |
ENSDART00000125577
|
taar19a
|
trace amine associated receptor 19a |
| chr7_+_17485782 | 0.11 |
ENSDART00000098083
|
CU179759.1
|
Danio rerio novel immune-type receptor 1d (nitr1d), mRNA. |
| chr3_+_24190207 | 0.11 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr18_+_33100606 | 0.10 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr6_+_42693114 | 0.10 |
ENSDART00000154353
|
si:ch211-207d10.2
|
si:ch211-207d10.2 |
| chr3_+_59411956 | 0.10 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr21_-_5879897 | 0.10 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr6_-_16981321 | 0.10 |
ENSDART00000154585
|
pimr29
|
Pim proto-oncogene, serine/threonine kinase, related 29 |
| chr6_+_16982613 | 0.10 |
ENSDART00000156104
|
pimr8
|
Pim proto-oncogene, serine/threonine kinase, related 8 |
| chr20_-_46046758 | 0.09 |
ENSDART00000127455
|
taar12j
|
trace amine associated receptor 12j |
| chr4_-_71177920 | 0.09 |
ENSDART00000158287
|
si:dkey-193i10.1
|
si:dkey-193i10.1 |
| chr12_-_44180132 | 0.09 |
ENSDART00000165998
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr24_-_25184553 | 0.08 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_-_65159258 | 0.08 |
ENSDART00000160429
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr16_-_21047872 | 0.08 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr8_-_14067517 | 0.07 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr15_-_46736432 | 0.07 |
ENSDART00000124567
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr18_+_45666489 | 0.07 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr1_+_45925150 | 0.06 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr3_+_49021079 | 0.06 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr21_-_35419486 | 0.06 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr4_-_17725008 | 0.06 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr4_-_36476889 | 0.04 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr16_-_31791165 | 0.04 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr9_-_40014339 | 0.04 |
ENSDART00000166918
|
IKZF2
|
si:zfos-1425h8.1 |
| chr14_+_18785727 | 0.04 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr23_-_44574059 | 0.03 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr18_+_7345417 | 0.03 |
ENSDART00000041429
|
glipr1b
|
GLI pathogenesis-related 1b |
| chr25_+_25438322 | 0.03 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr6_+_18142623 | 0.02 |
ENSDART00000169431
ENSDART00000158841 |
si:dkey-237i9.8
|
si:dkey-237i9.8 |
| chr3_+_16663373 | 0.01 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr5_-_15321451 | 0.01 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr15_-_766015 | 0.01 |
ENSDART00000190648
|
si:dkey-7i4.15
|
si:dkey-7i4.15 |
| chr21_-_35325466 | 0.00 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr18_-_21859019 | 0.00 |
ENSDART00000100885
|
nrn1la
|
neuritin 1-like a |
Network of associatons between targets according to the STRING database.
First level regulatory network of twist1a+twist1b+twist3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.3 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.1 | 0.4 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.2 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.2 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.2 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.2 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.2 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.4 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.2 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |