Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
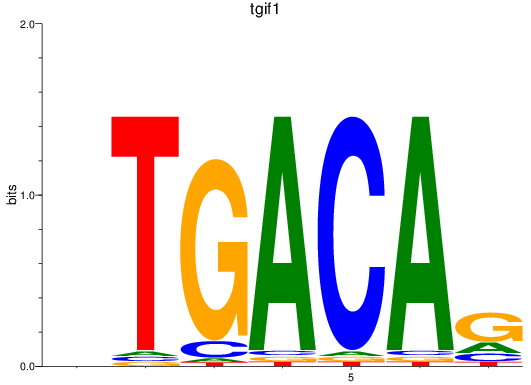
Results for tgif1
Z-value: 3.10
Transcription factors associated with tgif1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tgif1
|
ENSDARG00000059337 | TGFB-induced factor homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tgif1 | dr11_v1_chr24_-_9300160_9300160 | -0.95 | 2.2e-10 | Click! |
Activity profile of tgif1 motif
Sorted Z-values of tgif1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22272181 | 13.88 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr16_+_26012569 | 12.04 |
ENSDART00000148846
|
prss59.1
|
protease, serine, 59, tandem duplicate 1 |
| chr9_-_22232902 | 10.82 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr9_-_22129788 | 9.50 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr9_-_22205682 | 9.33 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr22_-_278328 | 8.60 |
ENSDART00000098072
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr20_-_25518488 | 8.58 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr3_-_1190132 | 8.43 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr9_-_22281854 | 8.02 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr16_-_19890303 | 7.49 |
ENSDART00000147161
ENSDART00000079159 |
hdac9b
|
histone deacetylase 9b |
| chr9_-_22135420 | 7.31 |
ENSDART00000184959
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr20_-_44576949 | 7.18 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr9_-_43071519 | 6.72 |
ENSDART00000109099
|
ttn.2
|
titin, tandem duplicate 2 |
| chr24_-_29997145 | 6.71 |
ENSDART00000135094
|
palmdb
|
palmdelphin b |
| chr9_+_42066030 | 6.57 |
ENSDART00000185311
ENSDART00000015267 |
pcbp3
|
poly(rC) binding protein 3 |
| chr6_-_9529217 | 6.43 |
ENSDART00000109113
|
si:ch211-113j14.1
|
si:ch211-113j14.1 |
| chr9_-_22117430 | 6.41 |
ENSDART00000101916
|
crygm2d15
|
crystallin, gamma M2d15 |
| chr9_-_22213297 | 6.39 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr5_+_45677781 | 6.32 |
ENSDART00000163120
ENSDART00000126537 |
gc
|
group-specific component (vitamin D binding protein) |
| chr19_-_44955710 | 6.07 |
ENSDART00000165246
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr6_-_13783604 | 6.04 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr9_-_22099536 | 6.01 |
ENSDART00000101923
|
CR391987.1
|
|
| chr18_-_5692292 | 5.80 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr20_+_35473288 | 5.73 |
ENSDART00000047195
|
mep1a.1
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 1 |
| chr19_-_12322356 | 5.69 |
ENSDART00000016128
|
ncaldb
|
neurocalcin delta b |
| chr13_+_28702104 | 5.67 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr19_+_43563179 | 5.65 |
ENSDART00000151478
|
cd164l2
|
CD164 sialomucin-like 2 |
| chr25_+_31227747 | 5.48 |
ENSDART00000033872
|
tnni2a.1
|
troponin I type 2a (skeletal, fast), tandem duplicate 1 |
| chr16_+_34528409 | 5.29 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr5_-_54197084 | 5.04 |
ENSDART00000163640
|
grk1b
|
G protein-coupled receptor kinase 1 b |
| chr11_+_1796426 | 5.03 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr8_-_25247284 | 5.02 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr1_-_43905252 | 4.98 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr3_-_40955780 | 4.91 |
ENSDART00000130130
|
cyp3c3
|
cytochrome P450, family 3, subfamily c, polypeptide 3 |
| chr16_+_5612547 | 4.91 |
ENSDART00000140226
ENSDART00000189352 |
CYTH2
|
si:dkey-283b15.4 |
| chr23_-_25976656 | 4.89 |
ENSDART00000019519
|
ada
|
adenosine deaminase |
| chr25_-_4482449 | 4.88 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr6_+_36381709 | 4.87 |
ENSDART00000004727
|
rhcgl1
|
Rh family, C glycoprotein, like 1 |
| chr23_-_1583193 | 4.80 |
ENSDART00000143841
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr21_+_25216397 | 4.78 |
ENSDART00000130970
|
sycn.3
|
syncollin, tandem duplicate 3 |
| chr23_-_30431333 | 4.69 |
ENSDART00000146633
|
camta1a
|
calmodulin binding transcription activator 1a |
| chr16_-_13789908 | 4.68 |
ENSDART00000138540
|
ttyh1
|
tweety family member 1 |
| chr12_-_4781801 | 4.66 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr13_+_45524475 | 4.66 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr14_-_2355833 | 4.59 |
ENSDART00000157677
|
PCDHGC3 (1 of many)
|
si:ch73-233f7.6 |
| chr3_-_28428198 | 4.55 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr21_-_14630831 | 4.48 |
ENSDART00000132142
ENSDART00000089967 |
cacna1bb
|
calcium channel, voltage-dependent, N type, alpha 1B subunit, b |
| chr6_-_27139396 | 4.40 |
ENSDART00000055848
|
zgc:103559
|
zgc:103559 |
| chr22_-_263117 | 4.40 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr2_-_12502495 | 4.33 |
ENSDART00000186781
|
dpp6b
|
dipeptidyl-peptidase 6b |
| chr23_+_40460333 | 4.32 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr24_-_24959607 | 4.32 |
ENSDART00000146930
ENSDART00000184482 |
pdk3a
|
pyruvate dehydrogenase kinase, isozyme 3a |
| chr15_-_2954443 | 4.30 |
ENSDART00000161053
|
zgc:153184
|
zgc:153184 |
| chr14_+_22172047 | 4.26 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr8_-_33114202 | 4.20 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr23_+_39854566 | 4.17 |
ENSDART00000190423
ENSDART00000164473 ENSDART00000161881 |
si:ch73-217b7.1
|
si:ch73-217b7.1 |
| chr21_+_40092301 | 4.06 |
ENSDART00000145150
|
serpinf2a
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2a |
| chr12_-_25916530 | 3.98 |
ENSDART00000186386
|
sncgb
|
synuclein, gamma b (breast cancer-specific protein 1) |
| chr4_-_815871 | 3.98 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr17_-_15149192 | 3.96 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr9_-_12493628 | 3.94 |
ENSDART00000180832
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr1_-_45553602 | 3.92 |
ENSDART00000143664
|
grin2bb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B, genome duplicate b |
| chr6_-_55585423 | 3.90 |
ENSDART00000157129
|
slc12a5a
|
solute carrier family 12 (potassium/chloride transporter), member 5a |
| chr1_-_7912932 | 3.90 |
ENSDART00000147383
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr24_-_5932982 | 3.89 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr23_+_19558574 | 3.89 |
ENSDART00000137811
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr11_-_9948487 | 3.88 |
ENSDART00000189677
ENSDART00000113171 |
nlgn1
|
neuroligin 1 |
| chr14_-_50892442 | 3.87 |
ENSDART00000174562
|
CDHR2
|
cadherin related family member 2 |
| chr23_+_39963599 | 3.87 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr25_+_33939728 | 3.84 |
ENSDART00000148537
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr7_-_20758825 | 3.81 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr13_-_23756700 | 3.81 |
ENSDART00000057612
|
rgs17
|
regulator of G protein signaling 17 |
| chr13_+_47821524 | 3.78 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr7_+_30371893 | 3.77 |
ENSDART00000075513
|
aqp9b
|
aquaporin 9b |
| chr17_+_15845765 | 3.76 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr8_+_19621511 | 3.76 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr14_-_36378494 | 3.72 |
ENSDART00000058503
|
gpm6aa
|
glycoprotein M6Aa |
| chr8_-_53195500 | 3.71 |
ENSDART00000132000
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr7_-_42206720 | 3.69 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr13_-_37519774 | 3.69 |
ENSDART00000141420
ENSDART00000185478 |
sgpp1
|
sphingosine-1-phosphate phosphatase 1 |
| chr10_+_36650222 | 3.68 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr5_-_67878064 | 3.64 |
ENSDART00000111203
|
tagln3a
|
transgelin 3a |
| chr2_+_20430366 | 3.59 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr16_-_12787029 | 3.59 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr16_-_16212615 | 3.58 |
ENSDART00000059905
|
upp1
|
uridine phosphorylase 1 |
| chr14_+_35691889 | 3.58 |
ENSDART00000074685
|
glrbb
|
glycine receptor, beta b |
| chr15_-_31067589 | 3.57 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr8_-_44835944 | 3.55 |
ENSDART00000185033
|
FO082780.1
|
|
| chr20_-_36393555 | 3.54 |
ENSDART00000153421
|
si:dkey-1j5.4
|
si:dkey-1j5.4 |
| chr17_+_25414033 | 3.53 |
ENSDART00000001691
|
tdh2
|
L-threonine dehydrogenase 2 |
| chr1_+_29096881 | 3.53 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr5_-_13766651 | 3.53 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr11_-_1131569 | 3.53 |
ENSDART00000173228
|
slc6a11b
|
solute carrier family 6 (neurotransmitter transporter), member 11b |
| chr5_+_6796291 | 3.51 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr7_+_73332486 | 3.49 |
ENSDART00000174119
ENSDART00000092051 ENSDART00000192388 |
CABZ01081780.1
|
|
| chr20_-_45708962 | 3.48 |
ENSDART00000124283
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr11_-_762721 | 3.47 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr12_-_22039350 | 3.40 |
ENSDART00000153187
|
thrab
|
thyroid hormone receptor alpha b |
| chr2_+_50999675 | 3.39 |
ENSDART00000158064
ENSDART00000165746 ENSDART00000163917 ENSDART00000172038 ENSDART00000169048 ENSDART00000164775 |
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr6_+_38381957 | 3.36 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr16_-_45178430 | 3.35 |
ENSDART00000165186
|
si:dkey-33i11.9
|
si:dkey-33i11.9 |
| chr1_-_37087966 | 3.34 |
ENSDART00000172111
ENSDART00000160056 |
nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr6_+_38427570 | 3.34 |
ENSDART00000170612
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr17_-_22067451 | 3.32 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr6_+_58915889 | 3.30 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr16_-_13730152 | 3.30 |
ENSDART00000138772
|
ttyh1
|
tweety family member 1 |
| chr21_-_28523548 | 3.26 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr12_+_27285994 | 3.25 |
ENSDART00000087204
|
dusp3a
|
dual specificity phosphatase 3a |
| chr14_-_6527010 | 3.25 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr6_+_3280939 | 3.24 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr7_-_51528661 | 3.23 |
ENSDART00000174263
|
nhsl2
|
NHS-like 2 |
| chr18_+_783936 | 3.22 |
ENSDART00000193357
|
rpp25b
|
ribonuclease P and MRP subunit p25, b |
| chr9_-_35334642 | 3.21 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr9_-_3653259 | 3.20 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr5_-_18046053 | 3.19 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr6_-_8580857 | 3.17 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr13_+_7292061 | 3.17 |
ENSDART00000179504
|
CABZ01072077.1
|
Danio rerio neuroblast differentiation-associated protein AHNAK-like (LOC795051), mRNA. |
| chr1_+_53321878 | 3.12 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr16_+_11558868 | 3.12 |
ENSDART00000112497
ENSDART00000180445 |
zgc:198329
|
zgc:198329 |
| chr1_-_54765262 | 3.12 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr20_+_50956369 | 3.11 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr15_+_47008307 | 3.10 |
ENSDART00000085387
|
ecel1
|
endothelin converting enzyme-like 1 |
| chr16_-_34477805 | 3.10 |
ENSDART00000136546
|
serinc2l
|
serine incorporator 2, like |
| chr9_+_23255410 | 3.09 |
ENSDART00000113241
ENSDART00000137231 |
tmem163a
|
transmembrane protein 163a |
| chr25_+_19238175 | 3.08 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr1_+_2112726 | 3.08 |
ENSDART00000131714
ENSDART00000138396 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr9_+_40939336 | 3.07 |
ENSDART00000100386
|
mstnb
|
myostatin b |
| chr1_+_23162124 | 3.06 |
ENSDART00000188428
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr10_-_41450367 | 3.05 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr8_+_49975160 | 3.05 |
ENSDART00000156403
ENSDART00000080135 |
gfpt1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr21_+_11385031 | 3.05 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr13_-_11644806 | 3.05 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr21_+_25643880 | 3.05 |
ENSDART00000192392
ENSDART00000145091 |
tmem151a
|
transmembrane protein 151A |
| chr17_-_21993620 | 3.04 |
ENSDART00000179905
ENSDART00000078873 |
slc22a7b.3
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 3 |
| chr12_-_4672708 | 3.03 |
ENSDART00000152659
ENSDART00000186076 |
si:ch211-255p10.4
|
si:ch211-255p10.4 |
| chr16_-_13613475 | 3.02 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr25_-_3549321 | 3.01 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr19_-_27261102 | 3.00 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr23_+_44741500 | 3.00 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr2_+_45484183 | 2.99 |
ENSDART00000183490
|
si:ch211-66k16.28
|
si:ch211-66k16.28 |
| chr17_-_17130942 | 2.98 |
ENSDART00000064241
|
nrxn3a
|
neurexin 3a |
| chr11_-_32723851 | 2.98 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr17_-_16965809 | 2.98 |
ENSDART00000153697
|
nrxn3a
|
neurexin 3a |
| chr17_-_15382704 | 2.97 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr3_+_16722014 | 2.97 |
ENSDART00000008711
|
gys1
|
glycogen synthase 1 (muscle) |
| chr24_-_38574631 | 2.95 |
ENSDART00000154539
|
slc17a7b
|
solute carrier family 17 (vesicular glutamate transporter), member 7b |
| chr8_+_8845932 | 2.95 |
ENSDART00000112028
|
si:ch211-180f4.1
|
si:ch211-180f4.1 |
| chr16_+_10776688 | 2.94 |
ENSDART00000161969
ENSDART00000172657 |
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr6_-_59381391 | 2.94 |
ENSDART00000157066
|
ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr9_-_12575776 | 2.93 |
ENSDART00000128931
ENSDART00000182695 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr15_-_29510074 | 2.93 |
ENSDART00000184070
|
BX324227.3
|
|
| chr1_+_40237276 | 2.93 |
ENSDART00000037553
|
faah2a
|
fatty acid amide hydrolase 2a |
| chr24_-_21512425 | 2.93 |
ENSDART00000184857
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr4_-_18436899 | 2.92 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr10_-_32524035 | 2.89 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr3_+_30257582 | 2.88 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr8_+_46418996 | 2.87 |
ENSDART00000144285
|
si:ch211-196g2.4
|
si:ch211-196g2.4 |
| chr7_+_42206847 | 2.86 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr25_+_15939275 | 2.84 |
ENSDART00000126641
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr1_-_47089818 | 2.83 |
ENSDART00000132378
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr21_+_45387903 | 2.81 |
ENSDART00000186253
ENSDART00000075432 |
jade2
|
jade family PHD finger 2 |
| chr9_+_54417141 | 2.80 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr7_-_20731078 | 2.79 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr7_-_58269812 | 2.79 |
ENSDART00000050077
|
sdcbp
|
syndecan binding protein (syntenin) |
| chr7_-_39378903 | 2.79 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr13_+_1182257 | 2.78 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr10_-_22845485 | 2.78 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr13_-_40141630 | 2.78 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr4_+_10017049 | 2.78 |
ENSDART00000144175
|
ccdc136b
|
coiled-coil domain containing 136b |
| chr15_-_30816370 | 2.78 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr21_-_43079161 | 2.77 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr5_+_28848870 | 2.76 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr17_-_51938663 | 2.76 |
ENSDART00000179784
|
ERG28
|
ergosterol biosynthesis 28 homolog |
| chr15_+_42767803 | 2.76 |
ENSDART00000164879
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr1_+_45217425 | 2.76 |
ENSDART00000179983
ENSDART00000074683 |
EVI5L
|
si:ch211-239f4.1 |
| chr3_+_45401472 | 2.75 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr2_-_24444838 | 2.75 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr18_+_13792490 | 2.74 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr3_+_10637330 | 2.74 |
ENSDART00000129257
|
tmem220
|
transmembrane protein 220 |
| chr25_+_32525131 | 2.74 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr16_-_50175069 | 2.73 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr11_-_4235811 | 2.71 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr15_-_4415917 | 2.70 |
ENSDART00000062874
|
atp1b3b
|
ATPase Na+/K+ transporting subunit beta 3b |
| chr6_+_41503854 | 2.69 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr9_-_43375205 | 2.66 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr11_-_43226255 | 2.65 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr8_+_19621731 | 2.65 |
ENSDART00000144667
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr16_-_13818061 | 2.64 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr5_+_28433547 | 2.64 |
ENSDART00000172218
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr23_-_30960506 | 2.64 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr24_+_29381946 | 2.64 |
ENSDART00000189551
|
ntng1a
|
netrin g1a |
| chr20_-_29420713 | 2.64 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr20_-_44575103 | 2.63 |
ENSDART00000192573
|
ubxn2a
|
UBX domain protein 2A |
| chr17_-_50331351 | 2.63 |
ENSDART00000149294
|
otofb
|
otoferlin b |
| chr15_-_23467750 | 2.62 |
ENSDART00000148804
|
pdzd3a
|
PDZ domain containing 3a |
| chr9_-_9348058 | 2.62 |
ENSDART00000132257
|
zgc:113337
|
zgc:113337 |
| chr11_-_33919309 | 2.61 |
ENSDART00000078202
|
phka2
|
phosphorylase kinase, alpha 2 (liver) |
| chr5_-_57635664 | 2.59 |
ENSDART00000074306
|
hspb2
|
heat shock protein, alpha-crystallin-related, b2 |
| chr15_+_37559570 | 2.59 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr8_-_18899427 | 2.59 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tgif1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 1.5 | 4.4 | GO:0000256 | allantoin catabolic process(GO:0000256) purine nucleobase catabolic process(GO:0006145) |
| 1.5 | 5.8 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) |
| 1.0 | 4.0 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.9 | 4.7 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.9 | 0.9 | GO:0021755 | eurydendroid cell differentiation(GO:0021755) |
| 0.9 | 4.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.8 | 9.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.8 | 3.3 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) |
| 0.8 | 3.3 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.8 | 13.8 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.8 | 5.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.8 | 3.9 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.8 | 3.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.7 | 2.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.7 | 3.6 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.7 | 2.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.7 | 2.1 | GO:0044108 | calcitriol biosynthetic process from calciol(GO:0036378) vitamin D biosynthetic process(GO:0042368) cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.7 | 2.1 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.7 | 2.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.7 | 8.7 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.6 | 2.5 | GO:0010896 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.6 | 3.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.6 | 4.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.6 | 80.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.6 | 4.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.5 | 3.3 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 2.1 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.5 | 9.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.5 | 3.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.5 | 5.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.5 | 2.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.5 | 2.0 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.5 | 4.7 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.5 | 8.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.5 | 3.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.5 | 3.2 | GO:0003222 | ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.5 | 1.8 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.5 | 1.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.4 | 8.5 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.4 | 1.8 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.4 | 2.2 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.4 | 4.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.4 | 4.8 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.4 | 2.2 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.4 | 1.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.4 | 4.7 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.4 | 3.0 | GO:0032371 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.4 | 1.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.4 | 1.2 | GO:0048917 | posterior lateral line ganglion development(GO:0048917) |
| 0.4 | 1.7 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.4 | 2.1 | GO:0006212 | thymine catabolic process(GO:0006210) uracil catabolic process(GO:0006212) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.4 | 3.3 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.4 | 0.8 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.4 | 2.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.4 | 4.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.4 | 2.8 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.4 | 1.6 | GO:0070131 | regulation of mitochondrial translation(GO:0070129) positive regulation of mitochondrial translation(GO:0070131) |
| 0.4 | 3.5 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.4 | 2.3 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.4 | 2.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.4 | 1.8 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.4 | 5.8 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 1.4 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.4 | 1.1 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.4 | 2.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.4 | 3.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.4 | 3.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.3 | 1.4 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.3 | 1.0 | GO:0070228 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.3 | 1.7 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.3 | 1.0 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.3 | 3.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.3 | 1.7 | GO:0043901 | negative regulation of multi-organism process(GO:0043901) |
| 0.3 | 5.1 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.3 | 1.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 4.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.3 | 8.6 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 1.0 | GO:0097401 | regulation of cellular pH reduction(GO:0032847) synaptic vesicle lumen acidification(GO:0097401) regulation of synaptic vesicle lumen acidification(GO:1901546) |
| 0.3 | 1.0 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.3 | 1.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.3 | 2.2 | GO:0042311 | vasodilation(GO:0042311) |
| 0.3 | 1.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 1.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.3 | 1.9 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 2.8 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 0.9 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.3 | 1.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.3 | 1.8 | GO:0043584 | nose development(GO:0043584) |
| 0.3 | 24.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.3 | 1.2 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.3 | 0.3 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.3 | 0.9 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.3 | 2.6 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.3 | 4.4 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 4.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 4.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.3 | 2.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.3 | 2.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.3 | 2.9 | GO:0015810 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.3 | 1.4 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.3 | 1.7 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.3 | 0.3 | GO:0048016 | inositol phosphate-mediated signaling(GO:0048016) |
| 0.3 | 2.8 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 1.9 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.3 | 0.8 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 9.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.3 | 1.8 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.3 | 1.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.5 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.3 | 7.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.3 | 1.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.2 | 4.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.2 | 0.7 | GO:0086013 | membrane repolarization(GO:0086009) membrane repolarization during action potential(GO:0086011) membrane repolarization during cardiac muscle cell action potential(GO:0086013) cardiac muscle cell membrane repolarization(GO:0099622) |
| 0.2 | 7.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.2 | 1.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 1.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 3.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 0.9 | GO:0042148 | strand invasion(GO:0042148) |
| 0.2 | 3.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 5.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.2 | 1.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.2 | 1.4 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.2 | 3.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.2 | 1.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.2 | 1.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.2 | 11.9 | GO:0006821 | chloride transport(GO:0006821) |
| 0.2 | 0.9 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 2.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.2 | 1.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.3 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 | 0.6 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 1.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 7.8 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.2 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 1.0 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.2 | 2.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.2 | 1.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.0 | GO:0051705 | regulation of proton transport(GO:0010155) social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) multi-organism behavior(GO:0051705) |
| 0.2 | 1.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 4.1 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 | 1.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.2 | 0.9 | GO:1901910 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 1.1 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) |
| 0.2 | 8.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.4 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.2 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.2 | 2.8 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 1.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.9 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.2 | 1.5 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 3.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 1.5 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 1.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.2 | 0.5 | GO:1902803 | regulation of synaptic vesicle transport(GO:1902803) |
| 0.2 | 8.6 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 0.8 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.8 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.2 | 0.5 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.2 | 3.2 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.2 | 1.3 | GO:0032411 | positive regulation of transporter activity(GO:0032411) |
| 0.2 | 1.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 0.5 | GO:0003218 | cardiac left ventricle morphogenesis(GO:0003214) cardiac left ventricle formation(GO:0003218) |
| 0.2 | 0.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.2 | 0.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 3.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.9 | GO:0003417 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.2 | 3.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 1.4 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) regulation of response to oxidative stress(GO:1902882) |
| 0.2 | 2.1 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 5.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 7.8 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.3 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 2.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.9 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 4.3 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 1.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 2.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.8 | GO:0034694 | response to prostaglandin(GO:0034694) cellular response to prostaglandin stimulus(GO:0071379) cellular response to fatty acid(GO:0071398) |
| 0.1 | 9.8 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 2.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.8 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 5.1 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.1 | 0.5 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.9 | GO:0070207 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.1 | 2.9 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 1.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.1 | 0.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.8 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 4.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.5 | GO:0090155 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.2 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.7 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 0.4 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 1.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 2.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 1.3 | GO:0046386 | deoxyribonucleotide catabolic process(GO:0009264) deoxyribose phosphate catabolic process(GO:0046386) |
| 0.1 | 10.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 2.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 2.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 1.9 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 6.4 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 2.6 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.4 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.5 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.7 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 2.5 | GO:0021986 | habenula development(GO:0021986) |
| 0.1 | 1.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 2.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.0 | GO:0042537 | benzene-containing compound metabolic process(GO:0042537) |
| 0.1 | 0.6 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.4 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.1 | 1.1 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.5 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.6 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 1.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.9 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 1.7 | GO:0071875 | adrenergic receptor signaling pathway(GO:0071875) |
| 0.1 | 0.6 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.1 | 0.7 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.8 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.1 | 1.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 5.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.1 | 1.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0050930 | regulation of positive chemotaxis(GO:0050926) positive regulation of positive chemotaxis(GO:0050927) induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 4.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.2 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 1.2 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.1 | 2.3 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.1 | 1.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.2 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 2.5 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.1 | 1.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 2.9 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 4.4 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.1 | 0.4 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 1.0 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 9.8 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.1 | 1.4 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.6 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.7 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 1.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 2.8 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.6 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.1 | 0.4 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.5 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.1 | 1.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.7 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 0.9 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 5.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.1 | 3.7 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.1 | 4.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 0.9 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.1 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 3.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.7 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 8.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0043648 | dicarboxylic acid metabolic process(GO:0043648) |
| 0.0 | 3.2 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 4.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.2 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.4 | GO:0046425 | regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.0 | 0.9 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 1.1 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 1.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.5 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 2.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 1.1 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.1 | GO:0051099 | positive regulation of DNA binding(GO:0043388) positive regulation of binding(GO:0051099) |
| 0.0 | 0.4 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:1990791 | central nervous system vasculogenesis(GO:0022009) dorsal root ganglion development(GO:1990791) |
| 0.0 | 1.4 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.4 | GO:0006032 | chitin catabolic process(GO:0006032) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 4.5 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.4 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 1.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0060606 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.2 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 1.5 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 2.1 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0033135 | regulation of peptidyl-serine phosphorylation(GO:0033135) positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.7 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.6 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 2.1 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.4 | GO:0015858 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.0 | 0.5 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 7.7 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.7 | GO:0033500 | carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) |
| 0.0 | 0.8 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 2.9 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 1.7 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.8 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.6 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 0.8 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.7 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.0 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.8 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 0.2 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 1.6 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 1.0 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.7 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.1 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.5 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 2.8 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.6 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.4 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0045576 | mast cell activation(GO:0045576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 3.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 1.5 | 10.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.8 | 9.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.8 | 2.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.6 | 3.9 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.6 | 8.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.6 | 1.7 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.6 | 4.6 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.6 | 5.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.5 | 2.7 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 1.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.5 | 2.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.5 | 16.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.5 | 3.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.5 | 3.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.4 | 4.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.4 | 6.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.4 | 6.0 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.3 | 13.9 | GO:0043679 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.3 | 3.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 1.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.3 | 2.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.3 | 2.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 1.7 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.3 | 2.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 1.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.0 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 4.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 5.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.2 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 5.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 0.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 1.7 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.2 | 24.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 2.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.2 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 0.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.2 | 9.9 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 8.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.9 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.2 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.5 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.2 | 2.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.2 | 0.8 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.2 | 11.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 1.5 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 3.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.6 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 8.2 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 2.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 0.6 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 2.5 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.4 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 1.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 2.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 1.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.6 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 1.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.9 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 0.7 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.0 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 1.5 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 2.4 | GO:0035371 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.1 | 13.1 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.1 | 5.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 4.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 2.0 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.1 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 0.7 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 2.6 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 4.4 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 3.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 6.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 2.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 8.8 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 2.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 5.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.9 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 8.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 5.0 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.7 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 12.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.3 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 26.1 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 27.2 | GO:0044421 | extracellular region part(GO:0044421) |
| 0.0 | 0.4 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.0 | 1.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.3 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 2.1 | 6.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 1.3 | 4.0 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 1.2 | 3.5 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 1.1 | 4.4 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 1.0 | 86.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.0 | 3.0 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 1.0 | 5.0 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 1.0 | 3.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 1.0 | 8.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.9 | 3.8 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.9 | 2.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.9 | 3.7 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.9 | 3.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.9 | 3.5 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.9 | 2.6 | GO:0030251 | cyclase inhibitor activity(GO:0010852) guanylate cyclase inhibitor activity(GO:0030251) |
| 0.8 | 3.1 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.8 | 2.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.7 | 3.0 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.7 | 2.9 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.7 | 2.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.7 | 5.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.7 | 3.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.7 | 2.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) cholesterol 26-hydroxylase activity(GO:0031073) |
| 0.7 | 2.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.6 | 1.9 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.6 | 3.7 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.6 | 3.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.6 | 1.8 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.6 | 4.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 4.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.5 | 4.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.5 | 7.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.5 | 2.1 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.5 | 2.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 5.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.5 | 1.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.5 | 4.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 16.9 | GO:0016917 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.5 | 1.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.5 | 2.3 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.5 | 1.9 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.4 | 4.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 4.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 1.7 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.4 | 5.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.4 | 4.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 3.4 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.4 | 1.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.4 | 2.1 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.4 | 6.7 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 1.2 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.4 | 1.8 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.4 | 3.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 3.4 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.3 | 2.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.3 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.3 | 4.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.3 | 4.0 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.3 | 4.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.3 | 1.3 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.3 | 3.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.3 | 5.8 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) |
| 0.3 | 2.3 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.3 | 6.4 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.3 | 1.6 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 0.9 | GO:0047325 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) |
| 0.3 | 2.2 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.3 | 0.9 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.3 | 4.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.3 | 2.7 | GO:0016933 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.3 | 3.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.3 | 2.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 1.5 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.8 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.3 | 0.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.3 | 8.1 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 2.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.3 | 9.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.3 | 1.6 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.3 | 2.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.3 | 1.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.3 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 2.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 4.9 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.2 | 4.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 6.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 2.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.2 | 4.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 0.9 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 1.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.2 | 2.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 3.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.4 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.2 | 0.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.2 | 3.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 2.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.8 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 1.5 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 2.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 5.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.2 | 0.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.2 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.8 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.2 | 3.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 3.7 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 0.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 1.3 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 4.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.8 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 0.9 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 13.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.2 | 1.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.5 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.2 | 1.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.2 | 1.8 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 5.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 6.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 4.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 8.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.4 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 2.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 1.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 3.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.6 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 3.1 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.8 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.1 | 1.5 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 2.1 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.5 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.8 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 14.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.9 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 6.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 1.0 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 2.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 11.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 1.3 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 3.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.1 | 2.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 1.0 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 1.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.5 | GO:0031690 | adrenergic receptor binding(GO:0031690) beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 1.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 5.2 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 2.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 2.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.0 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 2.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 11.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.6 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.6 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 3.3 | GO:0003823 | antigen binding(GO:0003823) |
| 0.1 | 2.7 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.1 | 1.7 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.1 | 2.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.6 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.7 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 23.0 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.3 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 1.7 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 1.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 2.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 3.8 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.2 | GO:1901612 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 20.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 2.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 2.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.6 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 5.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.1 | 0.8 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 5.5 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 0.3 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.1 | 2.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 2.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.1 | 8.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.5 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 1.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 6.8 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.1 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.4 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 1.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.0 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 8.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.7 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0034714 | type II transforming growth factor beta receptor binding(GO:0005114) type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 1.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.0 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 1.3 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 1.2 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.4 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.7 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 11.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.8 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 7.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 33.6 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.7 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.9 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.3 | GO:0052744 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 3.3 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.2 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 8.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:1990238 | double-stranded DNA endodeoxyribonuclease activity(GO:1990238) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.5 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.0 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 3.1 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.9 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 7.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.4 | 7.9 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 5.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.3 | 6.0 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.3 | 3.8 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 3.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 3.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.2 | 5.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 1.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.2 | 4.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 2.9 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 1.2 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 2.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 1.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 1.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.0 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 3.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 4.2 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.1 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 5.4 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.7 | 11.0 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.6 | 7.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.6 | 4.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.5 | 4.9 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.4 | 3.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.4 | 8.1 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.4 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 3.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.4 | 3.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.3 | 7.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.3 | 7.9 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.2 | 2.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 6.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.2 | 5.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 1.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.2 | 1.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.2 | 5.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 1.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.2 | 6.3 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.2 | 0.4 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 2.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 2.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 2.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.5 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 5.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 1.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 3.4 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 5.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.1 | 1.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.5 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 3.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.1 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 1.0 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.4 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 2.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |