Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
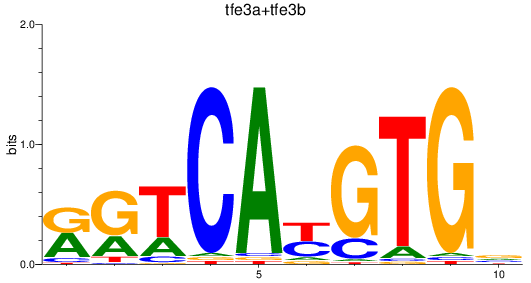
Results for tfe3a+tfe3b
Z-value: 0.70
Transcription factors associated with tfe3a+tfe3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfe3b
|
ENSDARG00000019457 | transcription factor binding to IGHM enhancer 3b |
|
tfe3a
|
ENSDARG00000098903 | transcription factor binding to IGHM enhancer 3a |
|
tfe3b
|
ENSDARG00000111231 | transcription factor binding to IGHM enhancer 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfe3b | dr11_v1_chr11_+_25508129_25508129 | 0.60 | 7.0e-03 | Click! |
| tfe3a | dr11_v1_chr8_+_7778770_7778770 | -0.10 | 7.0e-01 | Click! |
Activity profile of tfe3a+tfe3b motif
Sorted Z-values of tfe3a+tfe3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_4540127 | 1.23 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr18_+_26422124 | 1.13 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr17_+_32622933 | 1.12 |
ENSDART00000077418
|
ctsba
|
cathepsin Ba |
| chr1_+_10318089 | 1.11 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr5_+_61475451 | 1.10 |
ENSDART00000163444
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr19_+_15440841 | 0.99 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr9_-_14273652 | 0.95 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr19_+_15441022 | 0.92 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_-_24069331 | 0.89 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr8_-_50981175 | 0.84 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr15_-_43625549 | 0.82 |
ENSDART00000168589
|
ctsc
|
cathepsin C |
| chr15_+_1199407 | 0.81 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr6_+_59944488 | 0.77 |
ENSDART00000161158
|
nufip1
|
nuclear fragile X mental retardation protein interacting protein 1 |
| chr16_+_23960933 | 0.76 |
ENSDART00000146077
|
apoeb
|
apolipoprotein Eb |
| chr25_+_245438 | 0.75 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr16_-_9869056 | 0.75 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr10_+_10210455 | 0.72 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr23_-_31913231 | 0.71 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr21_-_25601648 | 0.71 |
ENSDART00000042578
|
efemp2b
|
EGF containing fibulin extracellular matrix protein 2b |
| chr24_-_42072886 | 0.71 |
ENSDART00000171389
|
CABZ01095370.1
|
|
| chr3_+_41922114 | 0.69 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr9_+_36946340 | 0.69 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr8_-_36554675 | 0.67 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr3_-_7296189 | 0.67 |
ENSDART00000148349
|
si:ch1073-186o8.3
|
si:ch1073-186o8.3 |
| chr3_+_23768898 | 0.67 |
ENSDART00000110682
|
hoxb1a
|
homeobox B1a |
| chr18_-_1414760 | 0.65 |
ENSDART00000171881
|
PEPD
|
peptidase D |
| chr21_-_39058490 | 0.64 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr13_+_22295905 | 0.64 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr3_-_49382896 | 0.64 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr13_+_13930263 | 0.63 |
ENSDART00000079154
|
rpia
|
ribose 5-phosphate isomerase A (ribose 5-phosphate epimerase) |
| chr17_+_32623931 | 0.63 |
ENSDART00000144217
|
ctsba
|
cathepsin Ba |
| chr19_-_26823647 | 0.62 |
ENSDART00000002464
|
neu1
|
neuraminidase 1 |
| chr19_-_1947403 | 0.61 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr22_+_30184039 | 0.60 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr19_-_42503143 | 0.60 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr25_+_17405458 | 0.58 |
ENSDART00000186711
|
e2f4
|
E2F transcription factor 4 |
| chr16_+_4654333 | 0.58 |
ENSDART00000167665
|
LIN28A
|
si:ch1073-284b18.2 |
| chr8_-_20118549 | 0.57 |
ENSDART00000132218
|
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_+_1579236 | 0.57 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr1_+_45754868 | 0.56 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr14_+_30543008 | 0.55 |
ENSDART00000145336
|
her5
|
hairy-related 5 |
| chr25_+_258883 | 0.55 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr20_-_16849306 | 0.55 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr15_+_1534644 | 0.55 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr7_+_65939098 | 0.54 |
ENSDART00000193599
|
RASSF10
|
Ras association domain family member 10 |
| chr22_+_18349794 | 0.54 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr24_-_12938922 | 0.53 |
ENSDART00000024084
|
pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr14_+_28442963 | 0.52 |
ENSDART00000186495
|
acsl4a
|
acyl-CoA synthetase long chain family member 4a |
| chr4_+_3358383 | 0.51 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr12_-_2800809 | 0.51 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr1_+_51827046 | 0.51 |
ENSDART00000052992
|
dand5
|
DAN domain family, member 5 |
| chr17_-_114121 | 0.51 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr17_+_37215820 | 0.51 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr21_-_19919918 | 0.50 |
ENSDART00000137307
ENSDART00000142523 ENSDART00000065670 |
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr21_-_45871866 | 0.50 |
ENSDART00000161716
|
larp1
|
La ribonucleoprotein domain family, member 1 |
| chr21_-_9914745 | 0.49 |
ENSDART00000172124
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr25_-_36369057 | 0.49 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr2_-_27575803 | 0.49 |
ENSDART00000014568
|
urod
|
uroporphyrinogen decarboxylase |
| chr21_-_27362938 | 0.49 |
ENSDART00000131297
|
rin1a
|
Ras and Rab interactor 1a |
| chr19_+_40641338 | 0.48 |
ENSDART00000146468
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr13_-_21650404 | 0.48 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr2_-_10877765 | 0.48 |
ENSDART00000100607
|
cdc7
|
cell division cycle 7 homolog (S. cerevisiae) |
| chr1_-_53504603 | 0.47 |
ENSDART00000162025
ENSDART00000100788 |
commd1
|
copper metabolism (Murr1) domain containing 1 |
| chr5_+_32847238 | 0.47 |
ENSDART00000144364
|
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr23_+_33947874 | 0.47 |
ENSDART00000136104
|
si:ch211-148l7.4
|
si:ch211-148l7.4 |
| chr8_-_16650595 | 0.47 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr19_-_22323695 | 0.46 |
ENSDART00000189355
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr12_+_7865470 | 0.46 |
ENSDART00000161683
|
BX548028.1
|
|
| chr7_+_55518519 | 0.46 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr21_+_8198652 | 0.46 |
ENSDART00000011096
|
nr6a1b
|
nuclear receptor subfamily 6, group A, member 1b |
| chr8_-_13735572 | 0.46 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr22_+_22302614 | 0.45 |
ENSDART00000049434
|
scamp4
|
secretory carrier membrane protein 4 |
| chr2_+_29976419 | 0.45 |
ENSDART00000056748
|
en2b
|
engrailed homeobox 2b |
| chr21_-_4539899 | 0.45 |
ENSDART00000112460
|
dolk
|
dolichol kinase |
| chr13_-_18122333 | 0.45 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr9_+_37152564 | 0.45 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr2_+_42005475 | 0.44 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr9_-_33063083 | 0.44 |
ENSDART00000048550
|
si:ch211-125e6.5
|
si:ch211-125e6.5 |
| chr25_+_34749187 | 0.44 |
ENSDART00000141473
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_-_50791280 | 0.44 |
ENSDART00000181224
|
CABZ01031870.1
|
|
| chr11_+_24819624 | 0.44 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr23_+_32028574 | 0.43 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr7_-_8961941 | 0.43 |
ENSDART00000111002
|
si:ch211-74f19.2
|
si:ch211-74f19.2 |
| chr6_+_59832786 | 0.43 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr2_-_43545342 | 0.43 |
ENSDART00000179796
|
CABZ01058721.1
|
|
| chr7_+_24390939 | 0.42 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr3_-_29962345 | 0.42 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr23_+_44461493 | 0.42 |
ENSDART00000149854
|
si:ch1073-228j22.1
|
si:ch1073-228j22.1 |
| chr16_+_23960744 | 0.42 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr25_+_17405201 | 0.42 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr23_+_42254960 | 0.42 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr1_+_604127 | 0.42 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr6_-_12644563 | 0.42 |
ENSDART00000153797
|
dock9b
|
dedicator of cytokinesis 9b |
| chr16_-_5143124 | 0.42 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr3_-_32362872 | 0.41 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr5_-_13167097 | 0.41 |
ENSDART00000149700
ENSDART00000030213 |
mapk1
|
mitogen-activated protein kinase 1 |
| chr6_+_45918981 | 0.41 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr4_+_5341592 | 0.41 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr5_+_4564233 | 0.41 |
ENSDART00000193435
|
CABZ01058647.1
|
|
| chr23_+_5565261 | 0.41 |
ENSDART00000059307
ENSDART00000169904 |
smpd2a
|
sphingomyelin phosphodiesterase 2a, neutral membrane (neutral sphingomyelinase) |
| chr1_+_19943803 | 0.41 |
ENSDART00000164661
|
apbb2b
|
amyloid beta (A4) precursor protein-binding, family B, member 2b |
| chr9_+_22782027 | 0.41 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr13_-_4992395 | 0.40 |
ENSDART00000102651
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr10_+_39283985 | 0.40 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr20_-_28698172 | 0.40 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr5_+_483965 | 0.40 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr24_-_21090447 | 0.40 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr20_+_25904199 | 0.39 |
ENSDART00000016864
|
slc35f6
|
solute carrier family 35, member F6 |
| chr25_+_31323978 | 0.39 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr8_-_16725573 | 0.39 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr12_-_19185865 | 0.39 |
ENSDART00000153343
|
zc3h7b
|
zinc finger CCCH-type containing 7B |
| chr7_+_46019780 | 0.38 |
ENSDART00000163991
|
ccne1
|
cyclin E1 |
| chr17_+_24590177 | 0.38 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr9_-_55586151 | 0.38 |
ENSDART00000181886
|
arsh
|
arylsulfatase H |
| chr2_-_14793343 | 0.38 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr25_-_35599887 | 0.38 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr11_-_4235811 | 0.38 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr19_+_167612 | 0.38 |
ENSDART00000169574
|
tatdn1
|
TatD DNase domain containing 1 |
| chr19_+_19737214 | 0.38 |
ENSDART00000160283
ENSDART00000169017 |
hoxa11a
|
homeobox A11a |
| chr1_+_19764995 | 0.37 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr17_-_45370200 | 0.37 |
ENSDART00000186208
|
znf106a
|
zinc finger protein 106a |
| chr3_-_25420931 | 0.37 |
ENSDART00000109601
ENSDART00000182184 |
bptf
|
bromodomain PHD finger transcription factor |
| chr11_-_31276064 | 0.37 |
ENSDART00000141062
ENSDART00000004780 |
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr22_-_11606078 | 0.36 |
ENSDART00000146026
|
pcyt2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr10_+_45148005 | 0.36 |
ENSDART00000182501
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr1_-_8141135 | 0.36 |
ENSDART00000152295
|
FAM83G
|
si:dkeyp-9d4.4 |
| chr19_-_2115040 | 0.36 |
ENSDART00000020497
|
snx13
|
sorting nexin 13 |
| chr10_-_105100 | 0.36 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr22_+_15979430 | 0.35 |
ENSDART00000189703
ENSDART00000192674 |
rc3h1a
|
ring finger and CCCH-type domains 1a |
| chr3_+_61391636 | 0.35 |
ENSDART00000126417
|
bri3
|
brain protein I3 |
| chr20_+_53474963 | 0.35 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr7_+_39410393 | 0.35 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr21_+_6394929 | 0.35 |
ENSDART00000138600
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr6_+_59642695 | 0.35 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr13_+_24662238 | 0.35 |
ENSDART00000014176
|
msx3
|
muscle segment homeobox 3 |
| chr9_+_12847040 | 0.34 |
ENSDART00000181983
ENSDART00000047191 |
glb1l
|
galactosidase, beta 1-like |
| chr23_-_19051710 | 0.34 |
ENSDART00000111852
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr22_+_9922301 | 0.34 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr18_-_16924221 | 0.34 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr17_-_44440832 | 0.33 |
ENSDART00000148786
|
exoc5
|
exocyst complex component 5 |
| chr1_-_25438934 | 0.33 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr7_-_40993456 | 0.33 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr15_-_14038227 | 0.33 |
ENSDART00000139068
|
zgc:114130
|
zgc:114130 |
| chr3_-_16784280 | 0.33 |
ENSDART00000137108
ENSDART00000137276 |
si:dkey-30j10.5
|
si:dkey-30j10.5 |
| chr12_-_3778848 | 0.33 |
ENSDART00000152128
|
si:ch211-166g5.4
|
si:ch211-166g5.4 |
| chr24_-_26854032 | 0.33 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr1_+_33322555 | 0.33 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr10_+_22510280 | 0.32 |
ENSDART00000109070
ENSDART00000182002 ENSDART00000192852 |
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr8_+_17168114 | 0.32 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr1_+_52462068 | 0.32 |
ENSDART00000124682
|
glb1
|
galactosidase, beta 1 |
| chr6_-_18976168 | 0.32 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| chr4_+_331351 | 0.32 |
ENSDART00000132625
|
tulp4a
|
tubby like protein 4a |
| chr23_-_31913069 | 0.32 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr24_-_16905018 | 0.32 |
ENSDART00000066759
|
mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr14_+_94946 | 0.32 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr6_-_2154137 | 0.31 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr19_+_19786117 | 0.31 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr13_+_24584401 | 0.31 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr16_+_14216581 | 0.31 |
ENSDART00000113093
|
gba
|
glucosidase, beta, acid |
| chr3_-_34296361 | 0.31 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr13_-_33114933 | 0.31 |
ENSDART00000140543
ENSDART00000075953 |
zfyve26
|
zinc finger, FYVE domain containing 26 |
| chr20_-_3911546 | 0.31 |
ENSDART00000169787
|
cnksr3
|
cnksr family member 3 |
| chr21_+_7298687 | 0.31 |
ENSDART00000187746
|
CU929160.1
|
|
| chr23_+_26079467 | 0.31 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr22_+_15343953 | 0.30 |
ENSDART00000045682
|
rrp36
|
ribosomal RNA processing 36 |
| chr19_+_34169055 | 0.30 |
ENSDART00000135592
ENSDART00000186043 |
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr8_-_16725959 | 0.30 |
ENSDART00000183593
|
depdc1a
|
DEP domain containing 1a |
| chr7_+_9189547 | 0.30 |
ENSDART00000169783
|
pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr25_-_19608382 | 0.30 |
ENSDART00000022279
ENSDART00000135201 ENSDART00000147223 ENSDART00000190220 ENSDART00000184242 ENSDART00000166824 |
gtse1
|
G-2 and S-phase expressed 1 |
| chr16_-_28268201 | 0.30 |
ENSDART00000121671
ENSDART00000141911 |
si:dkey-12j5.1
|
si:dkey-12j5.1 |
| chr18_+_8231138 | 0.30 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr5_-_2636078 | 0.30 |
ENSDART00000122274
|
cita
|
citron rho-interacting serine/threonine kinase a |
| chr23_-_19051869 | 0.30 |
ENSDART00000140866
|
arfgef2
|
ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) |
| chr5_-_22130937 | 0.30 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr4_-_49801268 | 0.30 |
ENSDART00000185457
|
si:ch211-197e7.1
|
si:ch211-197e7.1 |
| chr13_-_214122 | 0.30 |
ENSDART00000169273
|
ppp1r21
|
protein phosphatase 1, regulatory subunit 21 |
| chr13_+_28732101 | 0.30 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr5_-_54714789 | 0.30 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr25_+_22107643 | 0.30 |
ENSDART00000089680
|
sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr22_+_29991834 | 0.29 |
ENSDART00000147728
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr8_+_17529151 | 0.29 |
ENSDART00000147760
|
si:ch73-70k4.1
|
si:ch73-70k4.1 |
| chr9_+_24920677 | 0.29 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr12_+_34827808 | 0.29 |
ENSDART00000105533
|
tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr3_+_30500968 | 0.29 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr4_-_40972176 | 0.29 |
ENSDART00000152052
|
si:dkeyp-82h4.1
|
si:dkeyp-82h4.1 |
| chr22_-_10165446 | 0.29 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr25_-_25646806 | 0.29 |
ENSDART00000089066
|
tbc1d2b
|
TBC1 domain family, member 2B |
| chr15_-_4580763 | 0.29 |
ENSDART00000008170
|
rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr20_+_46213553 | 0.28 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr2_+_15128418 | 0.28 |
ENSDART00000141921
|
arhgap29b
|
Rho GTPase activating protein 29b |
| chr1_+_54737353 | 0.28 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr23_+_10146542 | 0.28 |
ENSDART00000048073
|
zgc:171775
|
zgc:171775 |
| chr7_-_19923249 | 0.28 |
ENSDART00000078694
|
zgc:110591
|
zgc:110591 |
| chr15_-_16177603 | 0.28 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr5_+_27404946 | 0.28 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr16_+_19029297 | 0.28 |
ENSDART00000115263
ENSDART00000114954 |
rapgef5b
|
Rap guanine nucleotide exchange factor (GEF) 5b |
| chr25_-_14424406 | 0.28 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr5_-_25072607 | 0.28 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr5_-_25066780 | 0.28 |
ENSDART00000002118
ENSDART00000182575 |
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr12_+_26538861 | 0.27 |
ENSDART00000152955
|
si:dkey-57h18.1
|
si:dkey-57h18.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfe3a+tfe3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 1.1 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 0.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.2 | 0.5 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 0.5 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.2 | 0.5 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.2 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.5 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 0.4 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.6 | GO:0042766 | nucleosome mobilization(GO:0042766) |
| 0.1 | 0.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 | 1.3 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.9 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.4 | GO:0010664 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.7 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.4 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.8 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.3 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.1 | 0.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 0.7 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.1 | 0.3 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.1 | 0.1 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.1 | 0.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.5 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.1 | 0.3 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.5 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.1 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.3 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.0 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.1 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.4 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.6 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.1 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.5 | GO:0033260 | nuclear DNA replication(GO:0033260) |
| 0.1 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.2 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 0.4 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.5 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.2 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.2 | GO:0035513 | oxidative RNA demethylation(GO:0035513) |
| 0.0 | 0.6 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.2 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.4 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.7 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:2001046 | positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 1.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.6 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 1.4 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 0.1 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.1 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.0 | 0.3 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.3 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 1.3 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.0 | 0.2 | GO:0051694 | skeletal muscle thin filament assembly(GO:0030240) pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.0 | 0.1 | GO:0032615 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.0 | 0.2 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.1 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.5 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 1.2 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 1.7 | GO:0090504 | epiboly(GO:0090504) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.4 | GO:0000469 | cleavage involved in rRNA processing(GO:0000469) |
| 0.0 | 0.1 | GO:0030323 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:2001044 | regulation of integrin-mediated signaling pathway(GO:2001044) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.3 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.1 | GO:1904398 | positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.2 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.4 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.6 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.0 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.4 | GO:0001895 | retina homeostasis(GO:0001895) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.4 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 0.5 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 1.6 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.1 | 0.2 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.6 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.2 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.0 | 0.2 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.3 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.4 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 7.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 0.1 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.4 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.1 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.3 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.0 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.7 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 1.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.6 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 1.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.7 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 0.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.6 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.4 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.3 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 0.6 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.3 | GO:0052834 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.3 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.2 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.6 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.2 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.1 | 0.1 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.6 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.4 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.4 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 0.2 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 1.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.4 | GO:0008296 | deoxyribonuclease I activity(GO:0004530) 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0038132 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.7 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 0.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.8 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 0.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 1.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.3 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 1.4 | GO:0042803 | protein homodimerization activity(GO:0042803) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.3 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 0.9 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.7 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.3 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.0 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.4 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 2.0 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |