Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
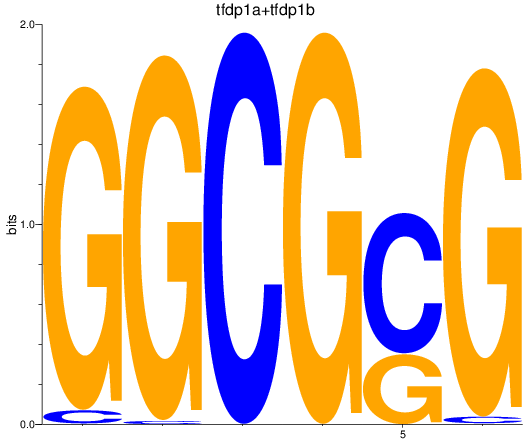
Results for tfdp1a+tfdp1b
Z-value: 3.35
Transcription factors associated with tfdp1a+tfdp1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfdp1b
|
ENSDARG00000016304 | transcription factor Dp-1, b |
|
tfdp1a
|
ENSDARG00000019293 | transcription factor Dp-1, a |
|
tfdp1a
|
ENSDARG00000111589 | transcription factor Dp-1, a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfdp1b | dr11_v1_chr1_+_230363_230363 | 0.90 | 1.9e-07 | Click! |
| tfdp1a | dr11_v1_chr9_+_34950942_34950942 | 0.84 | 5.7e-06 | Click! |
Activity profile of tfdp1a+tfdp1b motif
Sorted Z-values of tfdp1a+tfdp1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_5741733 | 12.07 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr8_+_29593986 | 10.68 |
ENSDART00000077642
|
atoh1a
|
atonal bHLH transcription factor 1a |
| chr23_-_3759345 | 9.93 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr19_-_31007417 | 9.79 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr23_-_3758637 | 7.94 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr8_-_2591654 | 7.64 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr7_+_49715750 | 7.01 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr23_-_23401305 | 6.77 |
ENSDART00000078936
|
her9
|
hairy-related 9 |
| chr3_+_41917499 | 6.17 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr13_+_46941930 | 6.13 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr20_+_2731436 | 6.09 |
ENSDART00000058779
ENSDART00000129870 ENSDART00000132186 ENSDART00000152727 |
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr4_+_14981854 | 5.78 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr24_+_27548474 | 5.72 |
ENSDART00000105774
ENSDART00000123368 |
ek1
|
eph-like kinase 1 |
| chr14_-_26704829 | 5.30 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr6_+_269204 | 5.27 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr25_+_3294150 | 5.08 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr8_+_387622 | 4.96 |
ENSDART00000167361
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr23_-_43718067 | 4.94 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr8_-_16259063 | 4.88 |
ENSDART00000057590
|
dmrta2
|
DMRT-like family A2 |
| chr25_+_28272908 | 4.80 |
ENSDART00000010325
|
fezf1
|
FEZ family zinc finger 1 |
| chr24_-_26995164 | 4.75 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr7_+_24881680 | 4.73 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr11_+_45153104 | 4.44 |
ENSDART00000159204
ENSDART00000177585 |
tk1
|
thymidine kinase 1, soluble |
| chr17_+_41463942 | 4.30 |
ENSDART00000075331
|
insm1b
|
insulinoma-associated 1b |
| chr23_+_4483083 | 4.29 |
ENSDART00000092389
|
nup210
|
nucleoporin 210 |
| chr4_+_17417111 | 4.18 |
ENSDART00000056005
|
ascl1a
|
achaete-scute family bHLH transcription factor 1a |
| chr24_+_41989108 | 4.17 |
ENSDART00000169725
|
zbtb14
|
zinc finger and BTB domain containing 14 |
| chr20_-_29498178 | 4.13 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr9_+_55857193 | 4.09 |
ENSDART00000160980
|
sept10
|
septin 10 |
| chr8_-_40327397 | 4.07 |
ENSDART00000074125
|
aplnra
|
apelin receptor a |
| chr25_+_20272145 | 4.00 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr10_-_76352 | 3.97 |
ENSDART00000186560
ENSDART00000144722 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr13_-_35908275 | 3.88 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr12_+_47663419 | 3.79 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr5_-_19400166 | 3.66 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr15_-_20233105 | 3.65 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr23_-_29751730 | 3.63 |
ENSDART00000056865
|
ctnnbip1
|
catenin, beta interacting protein 1 |
| chr4_+_306036 | 3.55 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr14_+_14841685 | 3.46 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr13_-_36844945 | 3.46 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr7_-_56766100 | 3.38 |
ENSDART00000189934
|
csnk2a2a
|
casein kinase 2, alpha prime polypeptide a |
| chr2_+_48303142 | 3.37 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr19_-_47570672 | 3.35 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr3_+_1211242 | 3.34 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr5_+_25760112 | 3.34 |
ENSDART00000088011
ENSDART00000182046 |
tmem2
|
transmembrane protein 2 |
| chr16_+_40576679 | 3.33 |
ENSDART00000169412
ENSDART00000193464 |
ccne2
|
cyclin E2 |
| chr3_+_23752150 | 3.29 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr13_+_9100 | 3.27 |
ENSDART00000165772
|
ppp4r3b
|
protein phosphatase 4, regulatory subunit 3B |
| chr12_-_31724198 | 3.26 |
ENSDART00000153056
ENSDART00000165299 ENSDART00000137464 ENSDART00000080173 |
srsf2a
|
serine/arginine-rich splicing factor 2a |
| chr17_+_132555 | 3.26 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr1_+_29858032 | 3.25 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr18_+_19975787 | 3.20 |
ENSDART00000138103
|
skor1b
|
SKI family transcriptional corepressor 1b |
| chr11_+_1551603 | 3.11 |
ENSDART00000185383
ENSDART00000121489 ENSDART00000040577 |
mybl2b
|
v-myb avian myeloblastosis viral oncogene homolog-like 2b |
| chr11_-_28224 | 3.11 |
ENSDART00000124104
|
sp1
|
sp1 transcription factor |
| chr6_-_33916756 | 3.10 |
ENSDART00000137447
ENSDART00000138488 |
nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chr7_-_48667056 | 3.09 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr12_+_26538861 | 3.06 |
ENSDART00000152955
|
si:dkey-57h18.1
|
si:dkey-57h18.1 |
| chr21_-_22357545 | 3.06 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr21_-_18275226 | 3.05 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr11_-_39202915 | 3.04 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr20_+_43648369 | 2.99 |
ENSDART00000187930
ENSDART00000017269 |
parp1
|
poly (ADP-ribose) polymerase 1 |
| chr22_-_24285432 | 2.97 |
ENSDART00000164083
|
si:ch211-117l17.4
|
si:ch211-117l17.4 |
| chr14_-_46374870 | 2.95 |
ENSDART00000185803
ENSDART00000188313 ENSDART00000031498 |
ccna2
|
cyclin A2 |
| chr16_-_55259199 | 2.94 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr25_+_36327034 | 2.92 |
ENSDART00000073452
|
zgc:110216
|
zgc:110216 |
| chr16_+_41015163 | 2.91 |
ENSDART00000058586
|
dek
|
DEK proto-oncogene |
| chr3_+_37574885 | 2.90 |
ENSDART00000055225
|
wnt9b
|
wingless-type MMTV integration site family, member 9B |
| chr7_-_26518086 | 2.90 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr11_+_24851671 | 2.90 |
ENSDART00000167659
|
ipo9
|
importin 9 |
| chr16_+_33953644 | 2.87 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr25_+_18556588 | 2.87 |
ENSDART00000073726
|
cav2
|
caveolin 2 |
| chr5_+_36895860 | 2.80 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr23_+_43718115 | 2.78 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr9_+_15890558 | 2.77 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr7_-_16597130 | 2.76 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr6_-_18618106 | 2.76 |
ENSDART00000161562
|
znf207b
|
zinc finger protein 207, b |
| chr25_-_6049339 | 2.76 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr5_+_68807170 | 2.76 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr18_+_62932 | 2.74 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr16_-_48430272 | 2.72 |
ENSDART00000005927
|
rad21a
|
RAD21 cohesin complex component a |
| chr5_+_22067570 | 2.72 |
ENSDART00000045574
|
shisa2a
|
shisa family member 2a |
| chr3_+_40409100 | 2.67 |
ENSDART00000103486
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr13_+_15933168 | 2.67 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr18_+_20034023 | 2.64 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr2_+_23701613 | 2.63 |
ENSDART00000047073
|
oxsr1a
|
oxidative stress responsive 1a |
| chr24_+_17005647 | 2.63 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr11_+_25403561 | 2.62 |
ENSDART00000089120
|
adnpa
|
activity-dependent neuroprotector homeobox a |
| chr8_+_27743550 | 2.62 |
ENSDART00000046004
|
wnt2bb
|
wingless-type MMTV integration site family, member 2Bb |
| chr24_+_24014880 | 2.61 |
ENSDART00000041335
|
chodl
|
chondrolectin |
| chr9_+_25775816 | 2.61 |
ENSDART00000127834
ENSDART00000189994 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr20_+_39344889 | 2.60 |
ENSDART00000009164
|
esco2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr6_+_36844127 | 2.60 |
ENSDART00000020505
|
chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr3_+_37790351 | 2.59 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr5_+_43870389 | 2.59 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr23_+_36178104 | 2.57 |
ENSDART00000103131
|
hoxc1a
|
homeobox C1a |
| chr20_+_1316495 | 2.54 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr21_+_18274825 | 2.53 |
ENSDART00000144322
ENSDART00000147768 |
wdr5
|
WD repeat domain 5 |
| chr1_+_19538299 | 2.52 |
ENSDART00000109416
|
smc2
|
structural maintenance of chromosomes 2 |
| chr17_+_8799451 | 2.52 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr20_+_1316803 | 2.52 |
ENSDART00000152586
ENSDART00000152165 |
nup43
|
nucleoporin 43 |
| chr1_-_21563040 | 2.52 |
ENSDART00000049572
|
ncapd3
|
non-SMC condensin II complex, subunit D3 |
| chr3_+_23743139 | 2.51 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr21_-_23307653 | 2.49 |
ENSDART00000140284
ENSDART00000134103 |
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr12_-_21684197 | 2.49 |
ENSDART00000152999
ENSDART00000153109 ENSDART00000148698 |
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr23_+_38245610 | 2.49 |
ENSDART00000191386
|
znf217
|
zinc finger protein 217 |
| chr20_-_48485354 | 2.46 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr9_-_48214216 | 2.45 |
ENSDART00000012938
|
phgdh
|
phosphoglycerate dehydrogenase |
| chr18_-_20458412 | 2.44 |
ENSDART00000012241
|
kif23
|
kinesin family member 23 |
| chr6_-_34220641 | 2.44 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr21_+_30721733 | 2.44 |
ENSDART00000040443
|
zgc:110224
|
zgc:110224 |
| chr22_-_7025393 | 2.44 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr7_-_16596938 | 2.43 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr22_+_16497670 | 2.43 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr18_-_20458840 | 2.43 |
ENSDART00000177125
|
kif23
|
kinesin family member 23 |
| chr12_+_10706772 | 2.41 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr17_+_8799661 | 2.41 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr10_-_29892486 | 2.41 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr12_+_45238292 | 2.41 |
ENSDART00000057983
|
mrpl38
|
mitochondrial ribosomal protein L38 |
| chr9_-_10068004 | 2.40 |
ENSDART00000011922
ENSDART00000162818 |
spopla
|
speckle-type POZ protein-like a |
| chr11_+_31324335 | 2.40 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr16_-_53919115 | 2.39 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| chr13_-_32906265 | 2.36 |
ENSDART00000113823
ENSDART00000171114 ENSDART00000180035 ENSDART00000137570 |
si:dkey-18j18.3
|
si:dkey-18j18.3 |
| chr14_-_246342 | 2.36 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr7_+_10911396 | 2.36 |
ENSDART00000167273
ENSDART00000081323 ENSDART00000170655 |
abhd17c
|
abhydrolase domain containing 17C |
| chr24_+_23791758 | 2.34 |
ENSDART00000066655
ENSDART00000146580 |
mybl1
|
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
| chr6_+_27090800 | 2.34 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr23_+_30898013 | 2.33 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr19_-_30811161 | 2.32 |
ENSDART00000103524
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr21_-_34658266 | 2.30 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr1_+_59538755 | 2.30 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr14_-_22113600 | 2.30 |
ENSDART00000113752
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr6_+_296130 | 2.30 |
ENSDART00000073985
|
rbfox2
|
RNA binding fox-1 homolog 2 |
| chr21_-_226071 | 2.30 |
ENSDART00000160667
|
nup54
|
nucleoporin 54 |
| chr19_+_7154500 | 2.28 |
ENSDART00000035967
ENSDART00000160894 |
brd2a
|
bromodomain containing 2a |
| chr12_+_9499993 | 2.28 |
ENSDART00000135871
|
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr15_-_16384184 | 2.27 |
ENSDART00000154504
|
fam222bb
|
family with sequence similarity 222, member Bb |
| chr4_-_7869731 | 2.21 |
ENSDART00000067339
|
mcm10
|
minichromosome maintenance 10 replication initiation factor |
| chr2_-_32666174 | 2.21 |
ENSDART00000133660
|
puf60a
|
poly-U binding splicing factor a |
| chr5_-_41307550 | 2.20 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr11_-_3907937 | 2.19 |
ENSDART00000082409
|
rpn1
|
ribophorin I |
| chr3_+_23742868 | 2.16 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr13_-_32899322 | 2.16 |
ENSDART00000133882
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr7_-_64971839 | 2.16 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr8_-_53960349 | 2.15 |
ENSDART00000160074
|
cdk11b
|
cyclin-dependent kinase 11B |
| chr22_-_3152357 | 2.15 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr13_-_32898962 | 2.14 |
ENSDART00000163757
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr5_+_36513605 | 2.14 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr20_+_33994580 | 2.14 |
ENSDART00000061729
|
si:dkey-97o5.1
|
si:dkey-97o5.1 |
| chr2_+_13907452 | 2.13 |
ENSDART00000169724
ENSDART00000190691 |
zgc:66475
|
zgc:66475 |
| chr1_+_19094548 | 2.12 |
ENSDART00000114514
|
ptpn9b
|
protein tyrosine phosphatase, non-receptor type 9, b |
| chr24_-_14577544 | 2.12 |
ENSDART00000066722
ENSDART00000109088 |
pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr24_-_13349464 | 2.12 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr2_-_26590628 | 2.10 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr6_-_22146258 | 2.10 |
ENSDART00000181700
|
gga1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr2_-_24962002 | 2.09 |
ENSDART00000132050
|
hltf
|
helicase-like transcription factor |
| chr12_+_11080776 | 2.06 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr19_-_17210760 | 2.06 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr11_-_40681011 | 2.06 |
ENSDART00000166372
ENSDART00000110622 ENSDART00000159713 |
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr20_-_25626693 | 2.06 |
ENSDART00000132247
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr8_+_23355484 | 2.05 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr12_+_5102670 | 2.05 |
ENSDART00000166600
|
cep55l
|
centrosomal protein 55 like |
| chr1_-_59313465 | 2.05 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr3_+_23697997 | 2.05 |
ENSDART00000184299
ENSDART00000078466 |
hoxb3a
|
homeobox B3a |
| chr5_+_2002804 | 2.05 |
ENSDART00000064088
|
vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr19_+_16064439 | 2.04 |
ENSDART00000151169
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr14_-_31618243 | 2.03 |
ENSDART00000016592
|
mmgt1
|
membrane magnesium transporter 1 |
| chr9_+_21401189 | 2.01 |
ENSDART00000062669
|
cx30.3
|
connexin 30.3 |
| chr13_-_29980215 | 2.00 |
ENSDART00000042049
|
hif1an
|
hypoxia inducible factor 1, alpha subunit inhibitor |
| chr9_-_21268576 | 1.99 |
ENSDART00000080604
|
sap18
|
Sin3A-associated protein |
| chr23_-_28347039 | 1.99 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr5_-_24869213 | 1.99 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr3_-_60602432 | 1.99 |
ENSDART00000163235
|
srsf2b
|
serine/arginine-rich splicing factor 2b |
| chr17_-_27419319 | 1.99 |
ENSDART00000127043
|
ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr5_-_1076431 | 1.97 |
ENSDART00000168336
|
si:zfos-128g4.1
|
si:zfos-128g4.1 |
| chr2_-_9744081 | 1.97 |
ENSDART00000097732
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr21_-_26490186 | 1.97 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr4_-_211714 | 1.97 |
ENSDART00000172566
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr15_+_19900197 | 1.96 |
ENSDART00000005221
|
thap12b
|
THAP domain containing 12b |
| chr12_+_34732262 | 1.96 |
ENSDART00000169634
|
si:dkey-21c1.8
|
si:dkey-21c1.8 |
| chr25_-_13403726 | 1.95 |
ENSDART00000056723
|
gins3
|
GINS complex subunit 3 |
| chr8_+_49766338 | 1.95 |
ENSDART00000060657
|
rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr13_-_4979029 | 1.95 |
ENSDART00000132931
|
nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr9_+_10692905 | 1.94 |
ENSDART00000061499
|
cxcr4b
|
chemokine (C-X-C motif), receptor 4b |
| chr4_-_9173552 | 1.94 |
ENSDART00000042963
|
chst11
|
carbohydrate (chondroitin 4) sulfotransferase 11 |
| chr5_+_34997763 | 1.94 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr13_+_14976108 | 1.92 |
ENSDART00000011520
|
noto
|
notochord homeobox |
| chr12_-_34716037 | 1.92 |
ENSDART00000152876
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr18_-_46010 | 1.92 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr10_-_44924289 | 1.92 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr10_+_10728870 | 1.91 |
ENSDART00000109282
|
swi5
|
SWI5 homologous recombination repair protein |
| chr6_-_137149 | 1.91 |
ENSDART00000186561
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr19_+_42432625 | 1.91 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr1_+_21563311 | 1.90 |
ENSDART00000147076
ENSDART00000006147 |
primpol
|
primase and polymerase (DNA-directed) |
| chr10_-_28835771 | 1.90 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr24_-_14577225 | 1.90 |
ENSDART00000135426
|
pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr24_+_11908480 | 1.89 |
ENSDART00000024224
|
fen1
|
flap structure-specific endonuclease 1 |
| chr1_-_27014872 | 1.89 |
ENSDART00000147414
ENSDART00000134032 ENSDART00000192087 ENSDART00000189111 ENSDART00000187348 ENSDART00000187248 |
cntln
|
centlein, centrosomal protein |
| chr22_+_2183110 | 1.89 |
ENSDART00000159279
ENSDART00000121703 |
znf1152
|
zinc finger protein 1152 |
| chr6_+_18544791 | 1.88 |
ENSDART00000167463
ENSDART00000169599 |
atad5b
|
ATPase family, AAA domain containing 5b |
| chr5_-_38121612 | 1.86 |
ENSDART00000159543
|
si:ch211-284e13.6
|
si:ch211-284e13.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfdp1a+tfdp1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.7 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 1.8 | 7.3 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 1.6 | 6.3 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 1.5 | 4.4 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.4 | 4.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 1.3 | 5.2 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 1.2 | 3.6 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 1.1 | 3.4 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 1.1 | 4.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 1.1 | 5.3 | GO:0048909 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 1.0 | 6.2 | GO:0055016 | hypochord development(GO:0055016) |
| 1.0 | 4.1 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 1.0 | 3.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 1.0 | 6.7 | GO:1902292 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.9 | 2.8 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.9 | 2.8 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.9 | 2.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.9 | 6.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.8 | 5.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.8 | 1.5 | GO:0009139 | pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) |
| 0.8 | 2.3 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.8 | 7.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.8 | 6.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.7 | 4.3 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.7 | 2.9 | GO:0071674 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.7 | 4.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.7 | 3.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.7 | 2.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.7 | 7.5 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.7 | 4.7 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.7 | 2.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.7 | 2.6 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.6 | 1.9 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.6 | 2.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 1.8 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.6 | 3.7 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.6 | 3.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.5 | 2.7 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.5 | 6.5 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 1.6 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.5 | 1.6 | GO:1903537 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.5 | 2.0 | GO:0010640 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.5 | 3.0 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.5 | 1.5 | GO:0050428 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.5 | 2.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.5 | 1.9 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.5 | 3.3 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.5 | 5.7 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.5 | 3.8 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.5 | 3.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.5 | 1.8 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.5 | 2.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.5 | 3.2 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.4 | 1.3 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.4 | 3.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.4 | 1.3 | GO:0030858 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.3 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.4 | 2.0 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.4 | 2.0 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.4 | 6.7 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.4 | 7.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.4 | 1.5 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.4 | 1.1 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.4 | 1.1 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.3 | 2.4 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.3 | 2.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 3.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.3 | 4.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.3 | 1.3 | GO:0072116 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.3 | 2.8 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.3 | 4.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.3 | 0.9 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.3 | 6.6 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.3 | 0.9 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.3 | 1.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 2.6 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.3 | 2.0 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.3 | 2.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.3 | 1.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 2.0 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 5.1 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.3 | 0.8 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.3 | 1.4 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 1.4 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.3 | 5.0 | GO:0006415 | translational termination(GO:0006415) |
| 0.3 | 1.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.3 | 1.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.3 | 2.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 1.3 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.3 | 2.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.2 | 3.0 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.2 | 1.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 6.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 2.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 1.6 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.2 | 0.7 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.2 | 2.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.2 | 0.9 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.2 | 0.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 4.0 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 1.3 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.2 | 1.5 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.2 | 2.8 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.2 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 0.6 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 11.4 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.2 | 1.0 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.2 | 0.6 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.2 | 1.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.2 | 1.0 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.2 | 1.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 18.9 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.2 | 2.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 0.8 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.2 | 0.9 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.2 | 1.1 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.2 | 2.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 1.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 1.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.2 | 0.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 2.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.2 | 2.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 9.9 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.2 | 6.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 3.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.2 | 1.7 | GO:0015800 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.2 | 2.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.2 | 3.4 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 1.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 | 0.5 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 0.6 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.5 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.2 | 1.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 2.0 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 9.6 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.2 | 0.5 | GO:0097384 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.1 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 1.3 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 2.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 5.1 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.1 | 1.8 | GO:0021754 | facial nucleus development(GO:0021754) commissural neuron axon guidance(GO:0071679) |
| 0.1 | 0.9 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 1.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.1 | 2.5 | GO:0006323 | DNA packaging(GO:0006323) |
| 0.1 | 4.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 2.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 2.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.2 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.1 | 0.5 | GO:0042779 | tRNA 3'-trailer cleavage(GO:0042779) |
| 0.1 | 0.5 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.5 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 5.9 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.5 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.1 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 1.9 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.5 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.9 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 1.5 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.1 | 0.4 | GO:1900158 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 0.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 3.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.7 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 15.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 5.4 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 2.4 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.7 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 1.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.7 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.3 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.7 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.6 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.1 | 2.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.6 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 1.6 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.1 | 2.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.8 | GO:0046070 | dGTP catabolic process(GO:0006203) purine nucleoside triphosphate catabolic process(GO:0009146) purine deoxyribonucleoside triphosphate catabolic process(GO:0009217) dGTP metabolic process(GO:0046070) |
| 0.1 | 0.5 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 2.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.3 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 5.2 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.1 | 1.8 | GO:0035476 | angioblast cell migration(GO:0035476) |
| 0.1 | 3.0 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.2 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 1.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 2.0 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 0.5 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.3 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.1 | 1.5 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.1 | 0.9 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 0.3 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.1 | 1.6 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.8 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 2.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.9 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.6 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 2.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.6 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 1.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 2.0 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.1 | 1.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.9 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.1 | 3.3 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.1 | 2.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.1 | 1.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 2.1 | GO:0021549 | cerebellum development(GO:0021549) |
| 0.1 | 5.0 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 0.6 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.2 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.9 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.1 | 2.4 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.1 | 1.1 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.9 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 0.6 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 15.3 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.1 | 4.3 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 1.0 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.1 | 1.0 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 5.3 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 3.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.7 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 | 1.9 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 0.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.4 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.7 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.1 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.8 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.8 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.8 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 2.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 4.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 1.8 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.5 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.0 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 1.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 1.8 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.6 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.9 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 1.4 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 2.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.6 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.8 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 1.2 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 2.4 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 1.3 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 6.1 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.9 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 1.8 | GO:0048593 | camera-type eye morphogenesis(GO:0048593) |
| 0.0 | 0.7 | GO:0098659 | inorganic cation import into cell(GO:0098659) inorganic ion import into cell(GO:0099587) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 3.7 | GO:0060070 | canonical Wnt signaling pathway(GO:0060070) |
| 0.0 | 5.1 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 1.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.9 | GO:0051090 | regulation of sequence-specific DNA binding transcription factor activity(GO:0051090) |
| 0.0 | 2.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.4 | GO:0007051 | spindle organization(GO:0007051) |
| 0.0 | 0.5 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.5 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.6 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 1.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 2.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.4 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.5 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 0.8 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.3 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 8.4 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.7 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 1.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 4.4 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 2.1 | GO:0014032 | neural crest cell development(GO:0014032) |
| 0.0 | 0.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.0 | 0.7 | GO:0030509 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.1 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.6 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 29.1 | GO:0006357 | regulation of transcription from RNA polymerase II promoter(GO:0006357) |
| 0.0 | 0.6 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.9 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.7 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.4 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.9 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 1.0 | 3.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.9 | 6.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.8 | 3.3 | GO:0097134 | cyclin E1-CDK2 complex(GO:0097134) |
| 0.8 | 2.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.8 | 3.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.7 | 6.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.7 | 3.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.6 | 6.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.6 | 2.3 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.6 | 8.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 3.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.5 | 2.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.5 | 6.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.5 | 5.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.4 | 1.8 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.4 | 3.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.4 | 2.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 2.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 5.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.4 | 1.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 2.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 2.7 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.4 | 1.5 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 1.9 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 3.3 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.4 | 1.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.4 | 2.9 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 3.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 5.2 | GO:0044545 | NSL complex(GO:0044545) |
| 0.3 | 1.0 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.3 | 2.0 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.3 | 1.6 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 7.0 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 6.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.3 | 7.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.3 | 4.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.3 | 3.5 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.3 | 10.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 1.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 5.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.3 | 3.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.3 | 2.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 0.7 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.2 | 2.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.6 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 0.2 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.2 | 1.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.2 | 2.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 2.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.2 | 8.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 2.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 1.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.2 | 1.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.2 | 16.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.2 | 2.7 | GO:0070461 | SAGA-type complex(GO:0070461) |
| 0.2 | 2.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 0.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 2.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 3.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.9 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.4 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 30.1 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 4.1 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 3.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 11.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 2.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 6.9 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 1.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 3.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 2.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.1 | 13.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 2.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 5.2 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 8.9 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 1.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 17.4 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.6 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 5.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.9 | GO:0045495 | pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 5.6 | GO:0031461 | cullin-RING ubiquitin ligase complex(GO:0031461) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.9 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 2.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 5.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 1.1 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 19.0 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.5 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 2.1 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.8 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.8 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 154.0 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) I band(GO:0031674) |
| 0.0 | 3.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 1.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.9 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.0 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 1.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.8 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.4 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 1.5 | 6.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 1.4 | 5.8 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 1.3 | 5.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 1.1 | 7.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.9 | 6.4 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.8 | 4.2 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.8 | 4.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.8 | 2.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.8 | 6.2 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.8 | 5.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.8 | 5.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.7 | 2.9 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.7 | 5.0 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.6 | 6.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.5 | 4.3 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.5 | 2.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.5 | 1.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.5 | 1.5 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.4 | 1.8 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.4 | 8.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.4 | 7.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.4 | 3.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 4.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.4 | 3.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 1.6 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 1.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 1.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.4 | 1.9 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.4 | 3.7 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 1.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.3 | 1.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 1.3 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.3 | 2.7 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.3 | 2.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.3 | 11.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.3 | 1.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.3 | 5.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.3 | 4.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 2.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.3 | 0.8 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.3 | 1.0 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.3 | 1.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.3 | 4.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 2.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.2 | 2.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.0 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.2 | 1.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 2.1 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.2 | 9.7 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.2 | 1.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.2 | 2.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.2 | 1.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.0 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 4.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.2 | 0.6 | GO:0000035 | acyl binding(GO:0000035) |
| 0.2 | 0.9 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.2 | 1.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 2.7 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.2 | 17.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 0.5 | GO:0042781 | 3'-tRNA processing endoribonuclease activity(GO:0042781) |
| 0.2 | 1.8 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 0.7 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.2 | 1.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 1.6 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.2 | 1.7 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.7 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 0.5 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 2.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.2 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 3.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 7.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.2 | 3.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.2 | 0.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 1.3 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 0.8 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 1.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 1.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.8 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.7 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.1 | 0.4 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 46.8 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 0.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 2.2 | GO:0061608 | nucleocytoplasmic transporter activity(GO:0005487) nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 2.2 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 7.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.6 | GO:0008565 | protein transmembrane transporter activity(GO:0008320) protein transporter activity(GO:0008565) |
| 0.1 | 1.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.8 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.9 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 1.1 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.9 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 4.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.8 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 2.0 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.6 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 2.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 2.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.4 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 5.3 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.1 | 5.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 0.8 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 10.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 2.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.6 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 1.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 0.9 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 1.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 3.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 4.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.1 | 2.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.9 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 24.4 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 36.0 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 0.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.9 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 4.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.1 | 3.0 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 2.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 1.9 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.7 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.4 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 69.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 1.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 2.7 | GO:0051540 | iron-sulfur cluster binding(GO:0051536) metal cluster binding(GO:0051540) |
| 0.0 | 0.9 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 3.4 | GO:0044389 | ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 1.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 39.1 | GO:0003677 | DNA binding(GO:0003677) |
| 0.0 | 0.9 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 4.0 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.7 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0008200 | ion channel inhibitor activity(GO:0008200) channel inhibitor activity(GO:0016248) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 2.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 3.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 13.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 3.5 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 4.1 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.4 | 32.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.3 | 4.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.3 | 9.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.3 | 4.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.2 | 5.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.2 | 2.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 4.6 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.2 | 7.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 3.0 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 3.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 6.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 7.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 3.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 9.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.9 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.0 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 0.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 1.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 1.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 3.5 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 1.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 4.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 3.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 2.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.9 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 2.0 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 2.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 2.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 0.9 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 0.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 0.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 0.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 1.1 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.5 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 15.5 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.8 | 10.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.7 | 22.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 7.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.4 | 4.6 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.4 | 5.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.4 | 1.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 6.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.4 | 3.3 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.3 | 13.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.3 | 3.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 3.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 4.0 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.3 | 8.7 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.3 | 5.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.3 | 4.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.2 | 5.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.2 | 4.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 3.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 2.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 3.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 2.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.2 | 2.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.2 | 1.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.2 | 1.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 0.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 0.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 0.8 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 7.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.8 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 5.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 1.8 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.7 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.0 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 2.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.9 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 0.5 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.1 | 0.8 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 0.8 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.9 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.9 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 1.9 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 3.9 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.9 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |