Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
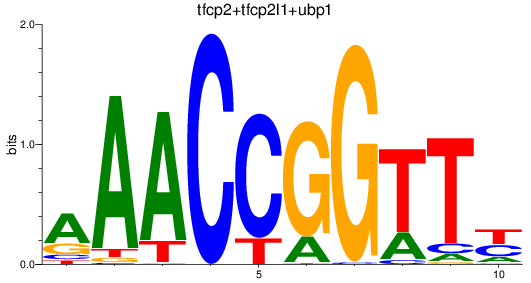
Results for tfcp2+tfcp2l1+ubp1
Z-value: 2.14
Transcription factors associated with tfcp2+tfcp2l1+ubp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ubp1
|
ENSDARG00000018000 | upstream binding protein 1 |
|
tfcp2l1
|
ENSDARG00000029497 | transcription factor CP2-like 1 |
|
tfcp2
|
ENSDARG00000060306 | transcription factor CP2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfcp2 | dr11_v1_chr23_-_33679579_33679579 | 0.74 | 2.6e-04 | Click! |
| tfcp2l1 | dr11_v1_chr9_+_38292947_38292947 | -0.70 | 8.7e-04 | Click! |
| ubp1 | dr11_v1_chr19_+_42806812_42806814 | -0.66 | 2.2e-03 | Click! |
Activity profile of tfcp2+tfcp2l1+ubp1 motif
Sorted Z-values of tfcp2+tfcp2l1+ubp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_18349794 | 7.66 |
ENSDART00000186580
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr17_+_51743908 | 7.07 |
ENSDART00000149039
ENSDART00000148869 |
odc1
|
ornithine decarboxylase 1 |
| chr8_-_13541514 | 6.38 |
ENSDART00000063834
|
zgc:86586
|
zgc:86586 |
| chr11_-_308838 | 6.01 |
ENSDART00000112538
|
poc1a
|
POC1 centriolar protein A |
| chr16_+_51180938 | 5.52 |
ENSDART00000169022
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr5_-_68795063 | 5.31 |
ENSDART00000016307
|
her1
|
hairy-related 1 |
| chr15_-_5901514 | 5.26 |
ENSDART00000155252
|
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr3_-_27647845 | 5.17 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr16_+_29509133 | 5.03 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr5_-_54712159 | 4.78 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr7_+_57088920 | 4.63 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr20_-_25631256 | 4.43 |
ENSDART00000048164
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr3_+_6469754 | 4.40 |
ENSDART00000185809
|
NUP85 (1 of many)
|
nucleoporin 85 |
| chr23_-_21446985 | 4.38 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr13_+_28495419 | 4.37 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr11_+_6116503 | 4.20 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr9_-_28939181 | 4.18 |
ENSDART00000101276
ENSDART00000135334 |
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_-_53685090 | 4.14 |
ENSDART00000162751
|
xpo1a
|
exportin 1 (CRM1 homolog, yeast) a |
| chr25_+_36292057 | 4.08 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr12_-_21681509 | 3.82 |
ENSDART00000112726
|
eme1
|
essential meiotic structure-specific endonuclease 1 |
| chr13_+_18533005 | 3.81 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr11_-_36350421 | 3.77 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr4_-_77979432 | 3.74 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr3_-_23512285 | 3.61 |
ENSDART00000159151
|
BX682558.1
|
|
| chr24_+_5935377 | 3.55 |
ENSDART00000191989
ENSDART00000185932 ENSDART00000131768 |
abi1a
|
abl-interactor 1a |
| chr15_+_5901970 | 3.55 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr16_-_27224246 | 3.53 |
ENSDART00000103257
|
alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr13_-_24260609 | 3.34 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr16_+_23961276 | 3.31 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr15_-_34668485 | 3.30 |
ENSDART00000186605
|
bag6
|
BCL2 associated athanogene 6 |
| chr4_+_10066840 | 3.29 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr22_+_18319230 | 3.28 |
ENSDART00000184747
ENSDART00000184649 |
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr7_+_57089354 | 3.19 |
ENSDART00000140702
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr6_+_41452979 | 3.19 |
ENSDART00000007353
|
wdr82
|
WD repeat domain 82 |
| chr4_+_28359718 | 3.16 |
ENSDART00000180048
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr4_+_18963822 | 3.12 |
ENSDART00000066975
ENSDART00000066973 |
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr13_+_24671481 | 3.09 |
ENSDART00000001678
|
adam8a
|
ADAM metallopeptidase domain 8a |
| chr11_-_36350876 | 3.02 |
ENSDART00000146495
ENSDART00000020655 |
psma5
|
proteasome subunit alpha 5 |
| chr17_-_19534474 | 3.02 |
ENSDART00000192469
|
cyp26c1
|
cytochrome P450, family 26, subfamily C, polypeptide 1 |
| chr21_+_25625026 | 3.00 |
ENSDART00000134678
|
ovol1b
|
ovo-like zinc finger 1b |
| chr19_-_47570672 | 2.99 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr7_-_26436436 | 2.96 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr7_+_25221757 | 2.94 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr22_+_19640309 | 2.93 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr8_-_32805214 | 2.93 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr13_-_25774183 | 2.91 |
ENSDART00000046981
|
pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr23_+_36144487 | 2.88 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr12_-_10476448 | 2.83 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr21_+_36396864 | 2.81 |
ENSDART00000137309
|
gemin5
|
gem (nuclear organelle) associated protein 5 |
| chr5_-_30704390 | 2.77 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr7_-_51648749 | 2.76 |
ENSDART00000109055
|
cited1
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 1 |
| chr21_+_19648814 | 2.75 |
ENSDART00000048581
|
fgf10a
|
fibroblast growth factor 10a |
| chr20_-_39367895 | 2.74 |
ENSDART00000136476
ENSDART00000021788 ENSDART00000180784 |
pbk
|
PDZ binding kinase |
| chr14_-_31087830 | 2.69 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr15_-_4596623 | 2.66 |
ENSDART00000132227
|
eif4h
|
eukaryotic translation initiation factor 4h |
| chr24_+_13925066 | 2.64 |
ENSDART00000134221
ENSDART00000012253 ENSDART00000081595 ENSDART00000136443 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr7_+_38808027 | 2.64 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr20_-_25645150 | 2.63 |
ENSDART00000063137
|
si:dkeyp-117h8.4
|
si:dkeyp-117h8.4 |
| chr19_-_17304866 | 2.62 |
ENSDART00000160433
ENSDART00000190798 ENSDART00000171284 |
sf3a3
|
splicing factor 3a, subunit 3 |
| chr19_-_18418763 | 2.61 |
ENSDART00000167271
|
zgc:112966
|
zgc:112966 |
| chr3_-_30488063 | 2.60 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr8_-_20914829 | 2.58 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr16_-_6424816 | 2.57 |
ENSDART00000164864
ENSDART00000141860 |
mboat1
|
membrane bound O-acyltransferase domain containing 1 |
| chr25_+_36292465 | 2.55 |
ENSDART00000152649
|
bmb
|
brambleberry |
| chr20_-_39391833 | 2.53 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr7_+_22792895 | 2.52 |
ENSDART00000184407
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr18_+_17725410 | 2.48 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr11_+_2202987 | 2.44 |
ENSDART00000190008
ENSDART00000173139 |
hoxc6b
|
homeobox C6b |
| chr17_+_26815021 | 2.43 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr1_-_11104805 | 2.42 |
ENSDART00000147648
|
knl1
|
kinetochore scaffold 1 |
| chr3_-_40162843 | 2.40 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr12_+_27024676 | 2.40 |
ENSDART00000153104
|
msl1b
|
male-specific lethal 1 homolog b (Drosophila) |
| chr5_+_20319519 | 2.39 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr4_-_26035770 | 2.39 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr4_+_6032640 | 2.38 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr7_-_17814118 | 2.38 |
ENSDART00000179688
|
ecsit
|
ECSIT signalling integrator |
| chr22_+_17499905 | 2.35 |
ENSDART00000061366
ENSDART00000133793 |
rab11ba
|
RAB11B, member RAS oncogene family, a |
| chr21_+_22892836 | 2.32 |
ENSDART00000065565
|
alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr14_-_33425170 | 2.31 |
ENSDART00000124629
ENSDART00000105800 ENSDART00000001318 |
nkap
|
NFKB activating protein |
| chr8_-_36554675 | 2.31 |
ENSDART00000132804
ENSDART00000078746 |
ccdc157
|
coiled-coil domain containing 157 |
| chr22_+_18319666 | 2.31 |
ENSDART00000033103
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr2_-_9527129 | 2.30 |
ENSDART00000157422
ENSDART00000004398 |
cope
|
coatomer protein complex, subunit epsilon |
| chr2_-_25143373 | 2.30 |
ENSDART00000160108
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr18_-_7448047 | 2.29 |
ENSDART00000193213
ENSDART00000131940 ENSDART00000186944 ENSDART00000052803 |
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr22_-_15587360 | 2.28 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr4_-_48636872 | 2.28 |
ENSDART00000168605
|
znf1063
|
zinc finger protein 1063 |
| chr22_-_3299355 | 2.21 |
ENSDART00000190993
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr3_-_26806032 | 2.21 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr9_-_3400727 | 2.16 |
ENSDART00000183979
ENSDART00000111386 |
dlx2a
|
distal-less homeobox 2a |
| chr12_+_27156943 | 2.15 |
ENSDART00000153030
ENSDART00000001737 |
skap1
|
src kinase associated phosphoprotein 1 |
| chr1_+_51475094 | 2.14 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr14_+_6535426 | 2.14 |
ENSDART00000055961
|
thg1l
|
tRNA-histidine guanylyltransferase 1-like |
| chr21_-_40049642 | 2.13 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr8_-_52715911 | 2.12 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr5_+_30741730 | 2.11 |
ENSDART00000098246
ENSDART00000186992 ENSDART00000182533 |
ftr83
|
finTRIM family, member 83 |
| chr10_+_44692272 | 2.11 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr9_-_45602978 | 2.11 |
ENSDART00000139019
ENSDART00000085763 |
agr1
|
anterior gradient 1 |
| chr11_+_3246059 | 2.11 |
ENSDART00000161529
|
timeless
|
timeless circadian clock |
| chr5_+_43807003 | 2.10 |
ENSDART00000097625
|
zgc:158640
|
zgc:158640 |
| chr2_+_53720028 | 2.10 |
ENSDART00000170799
|
ctnnbl1
|
catenin, beta like 1 |
| chr13_-_12006007 | 2.10 |
ENSDART00000111438
|
mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr9_-_10068004 | 2.09 |
ENSDART00000011922
ENSDART00000162818 |
spopla
|
speckle-type POZ protein-like a |
| chr9_+_21260314 | 2.09 |
ENSDART00000145943
|
ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr17_-_43677471 | 2.08 |
ENSDART00000137015
|
egr2a
|
early growth response 2a |
| chr19_-_42588510 | 2.08 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr3_-_29977495 | 2.07 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr14_+_52481288 | 2.05 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr14_-_31862657 | 2.05 |
ENSDART00000172870
ENSDART00000007927 ENSDART00000134748 ENSDART00000128730 |
rbmx
|
RNA binding motif protein, X-linked |
| chr6_-_27108844 | 2.04 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr21_-_36396334 | 2.04 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr16_+_33938227 | 2.02 |
ENSDART00000166254
|
gpn2
|
GPN-loop GTPase 2 |
| chr13_-_40411908 | 2.02 |
ENSDART00000057094
ENSDART00000150091 |
nkx2.3
|
NK2 homeobox 3 |
| chr13_-_15928934 | 2.01 |
ENSDART00000142732
|
ttl
|
tubulin tyrosine ligase |
| chr14_+_9421510 | 1.98 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr21_-_37973081 | 1.97 |
ENSDART00000136569
|
ripply1
|
ripply transcriptional repressor 1 |
| chr16_-_27224000 | 1.96 |
ENSDART00000126347
|
alg2
|
asparagine-linked glycosylation 2 (alpha-1,3-mannosyltransferase) |
| chr2_-_47620806 | 1.96 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr6_-_28943056 | 1.96 |
ENSDART00000065138
|
tbc1d23
|
TBC1 domain family, member 23 |
| chr6_+_149405 | 1.95 |
ENSDART00000161154
|
fdx1l
|
ferredoxin 1-like |
| chr20_-_22193190 | 1.95 |
ENSDART00000047624
|
tmem165
|
transmembrane protein 165 |
| chr23_-_25779995 | 1.93 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr21_-_13149453 | 1.92 |
ENSDART00000172578
|
si:dkey-228b2.6
|
si:dkey-228b2.6 |
| chr5_-_28041715 | 1.91 |
ENSDART00000078660
|
zgc:113436
|
zgc:113436 |
| chr18_+_5308392 | 1.90 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr8_-_3312384 | 1.88 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr4_+_53976731 | 1.87 |
ENSDART00000165813
|
si:ch211-249c2.1
|
si:ch211-249c2.1 |
| chr3_+_32341917 | 1.87 |
ENSDART00000055299
|
prl
|
prolactin |
| chr7_+_19835569 | 1.87 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr20_-_36416922 | 1.86 |
ENSDART00000019145
|
lbr
|
lamin B receptor |
| chr21_-_13668358 | 1.86 |
ENSDART00000180323
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr11_+_44579865 | 1.85 |
ENSDART00000173425
|
nid1b
|
nidogen 1b |
| chr3_-_26204867 | 1.85 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr6_+_23810529 | 1.85 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr18_-_40773413 | 1.84 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr8_-_28349859 | 1.84 |
ENSDART00000062671
|
tuba8l
|
tubulin, alpha 8 like |
| chr13_+_31321297 | 1.83 |
ENSDART00000143308
|
antxr1d
|
anthrax toxin receptor 1d |
| chr7_+_24205034 | 1.83 |
ENSDART00000018580
|
nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr12_-_7607114 | 1.83 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr9_-_41088279 | 1.81 |
ENSDART00000000564
|
asnsd1
|
asparagine synthetase domain containing 1 |
| chr10_-_1523253 | 1.80 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr9_-_28255029 | 1.80 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr22_-_17499513 | 1.80 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr24_+_38671054 | 1.79 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr2_-_42035250 | 1.77 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr5_+_15203421 | 1.76 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr10_+_36662640 | 1.74 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr3_-_7524363 | 1.74 |
ENSDART00000162970
|
znf1001
|
zinc finger protein 1001 |
| chr14_-_41467497 | 1.73 |
ENSDART00000181220
|
mid1ip1l
|
MID1 interacting protein 1, like |
| chr21_+_34088377 | 1.72 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr4_-_35989745 | 1.71 |
ENSDART00000162568
|
znf1125
|
zinc finger protein 1125 |
| chr25_+_7532627 | 1.69 |
ENSDART00000187660
|
ptdss2
|
phosphatidylserine synthase 2 |
| chr1_-_38195012 | 1.69 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr4_-_50434519 | 1.69 |
ENSDART00000150372
|
znf1061
|
zinc finger protein 1061 |
| chr22_+_22021936 | 1.69 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr10_+_39084354 | 1.67 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr2_+_327081 | 1.65 |
ENSDART00000155595
|
zgc:174263
|
zgc:174263 |
| chr3_-_36846496 | 1.63 |
ENSDART00000055237
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr3_-_38783951 | 1.63 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr19_+_9091673 | 1.63 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr4_-_64141714 | 1.60 |
ENSDART00000128628
|
BX914205.3
|
|
| chr2_-_30611389 | 1.59 |
ENSDART00000142500
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr15_+_46329149 | 1.58 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr6_+_13506841 | 1.57 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr21_-_32467799 | 1.57 |
ENSDART00000007675
ENSDART00000133099 |
zgc:123105
|
zgc:123105 |
| chr16_+_17252487 | 1.57 |
ENSDART00000063572
|
gtf3c6
|
general transcription factor IIIC, polypeptide 6, alpha |
| chr9_-_28939796 | 1.55 |
ENSDART00000101269
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr5_-_46329880 | 1.54 |
ENSDART00000156577
|
si:ch211-130m23.5
|
si:ch211-130m23.5 |
| chr4_-_27129697 | 1.52 |
ENSDART00000131240
|
zbed4
|
zinc finger, BED-type containing 4 |
| chr21_-_30031396 | 1.51 |
ENSDART00000157167
|
pwwp2a
|
PWWP domain containing 2A |
| chr7_-_34256374 | 1.50 |
ENSDART00000075176
|
dis3l
|
DIS3 like exosome 3'-5' exoribonuclease |
| chr25_-_34670413 | 1.50 |
ENSDART00000073440
|
DNAJA4
|
DnaJ heat shock protein family (Hsp40) member A4 |
| chr24_-_9979342 | 1.50 |
ENSDART00000138576
ENSDART00000191206 |
zgc:171977
|
zgc:171977 |
| chr1_-_46706639 | 1.49 |
ENSDART00000074519
|
kpna3
|
karyopherin alpha 3 (importin alpha 4) |
| chr24_+_20536056 | 1.48 |
ENSDART00000082082
|
gars
|
glycyl-tRNA synthetase |
| chr9_-_10778615 | 1.48 |
ENSDART00000182577
|
CABZ01053619.1
|
|
| chr7_+_48806420 | 1.47 |
ENSDART00000083431
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr11_-_309420 | 1.46 |
ENSDART00000173185
|
poc1a
|
POC1 centriolar protein A |
| chr20_-_14012859 | 1.45 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr21_-_32467099 | 1.44 |
ENSDART00000186354
|
zgc:123105
|
zgc:123105 |
| chr22_-_21176269 | 1.42 |
ENSDART00000112839
|
rex1bd
|
required for excision 1-B domain containing |
| chr15_-_34866219 | 1.42 |
ENSDART00000099723
|
sh3bp5la
|
SH3-binding domain protein 5-like, a |
| chr6_+_37625787 | 1.39 |
ENSDART00000065122
|
tubgcp5
|
tubulin, gamma complex associated protein 5 |
| chr6_+_52931841 | 1.39 |
ENSDART00000174358
|
si:dkeyp-3f10.12
|
si:dkeyp-3f10.12 |
| chr14_-_45558490 | 1.38 |
ENSDART00000165060
|
ints5
|
integrator complex subunit 5 |
| chr4_+_42078825 | 1.38 |
ENSDART00000164496
|
znf1060
|
zinc finger protein 1060 |
| chr22_-_34979139 | 1.38 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr24_+_37825634 | 1.36 |
ENSDART00000129889
|
ift140
|
intraflagellar transport 140 homolog (Chlamydomonas) |
| chr13_+_33268657 | 1.35 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr25_+_30196039 | 1.35 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr3_-_32590164 | 1.35 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr8_-_28274552 | 1.34 |
ENSDART00000131580
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr7_-_40656148 | 1.33 |
ENSDART00000142315
|
nom1
|
nucleolar protein with MIF4G domain 1 |
| chr13_-_36798204 | 1.33 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr22_-_14161309 | 1.33 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr2_+_47906240 | 1.33 |
ENSDART00000122206
|
ftr23
|
finTRIM family, member 23 |
| chr21_-_32082130 | 1.32 |
ENSDART00000003978
ENSDART00000182050 |
mat2b
|
methionine adenosyltransferase II, beta |
| chr21_-_32081552 | 1.32 |
ENSDART00000135659
|
mat2b
|
methionine adenosyltransferase II, beta |
| chr4_-_193762 | 1.31 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr20_+_26916639 | 1.31 |
ENSDART00000077787
|
serpinb1l2
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 2 |
| chr5_+_31779911 | 1.29 |
ENSDART00000098163
|
slc25a25b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25b |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfcp2+tfcp2l1+ubp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.6 | GO:0000741 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 1.1 | 3.3 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.1 | 3.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 1.0 | 3.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.8 | 7.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.8 | 2.4 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.8 | 7.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.7 | 5.7 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.7 | 2.7 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.7 | 2.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.7 | 2.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.6 | 1.9 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.6 | 1.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.6 | 2.4 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.6 | 1.8 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.5 | 4.8 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.5 | 7.8 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.5 | 1.9 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.5 | 4.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.5 | 1.8 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.4 | 4.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.4 | 2.1 | GO:0043111 | replication fork arrest(GO:0043111) |
| 0.4 | 2.0 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.4 | 2.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.4 | 3.8 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.4 | 2.7 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.4 | 2.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.4 | 3.4 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 2.2 | GO:0046532 | amacrine cell differentiation(GO:0035881) regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.4 | 1.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.4 | 1.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 2.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 3.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 3.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.3 | 2.3 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.3 | 1.9 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.3 | 3.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.3 | 2.6 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.3 | 1.7 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) determination of intestine left/right asymmetry(GO:0071908) |
| 0.3 | 7.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.3 | 1.3 | GO:0071320 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.3 | 1.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.3 | 1.6 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 0.2 | 1.0 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.2 | 0.7 | GO:0060958 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) endocardial cell development(GO:0060958) cell proliferation involved in heart valve development(GO:2000793) |
| 0.2 | 2.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.2 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 7.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.2 | 1.7 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.2 | 2.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 1.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.2 | 3.0 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.2 | 0.8 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.2 | 1.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.2 | 0.9 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 2.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 1.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.2 | 0.5 | GO:0030237 | female sex determination(GO:0030237) |
| 0.2 | 1.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 1.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.1 | 0.9 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 2.1 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.1 | 0.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.6 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 1.5 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.9 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.1 | 1.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.4 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 1.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.9 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 2.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 3.2 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 1.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 1.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.9 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.3 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.6 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.1 | 0.7 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.1 | 3.8 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 1.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 1.0 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.5 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.1 | 2.9 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 2.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 1.9 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.1 | 0.5 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 2.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.7 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 2.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 2.2 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.1 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) |
| 0.1 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 2.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.4 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.7 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.7 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 18.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.1 | 1.6 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 1.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.4 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.1 | 1.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 3.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.8 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.8 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 1.4 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) SMAD protein signal transduction(GO:0060395) |
| 0.0 | 1.6 | GO:0031623 | receptor internalization(GO:0031623) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 2.1 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 4.4 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 2.1 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 1.3 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 2.0 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 3.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 2.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0038066 | p38MAPK cascade(GO:0038066) regulation of p38MAPK cascade(GO:1900744) |
| 0.0 | 1.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 2.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 5.7 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 2.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 1.2 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 1.3 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 0.0 | 2.4 | GO:0009913 | epidermal cell differentiation(GO:0009913) |
| 0.0 | 2.1 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane(GO:0006620) posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.2 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 2.9 | GO:0071773 | BMP signaling pathway(GO:0030509) response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.8 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.4 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 10.3 | GO:0045184 | establishment of protein localization(GO:0045184) |
| 0.0 | 3.2 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.0 | 3.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.8 | 3.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.7 | 13.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.7 | 2.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.6 | 3.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.6 | 6.8 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.5 | 3.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.4 | 2.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.4 | 7.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 2.0 | GO:0071203 | WASH complex(GO:0071203) |
| 0.4 | 2.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 1.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.3 | 2.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 2.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.3 | 0.9 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 2.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 2.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.3 | 2.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.3 | 0.9 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.3 | 1.6 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 2.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 3.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.2 | 1.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.2 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.2 | 1.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 2.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.5 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 2.0 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.1 | 2.9 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 0.5 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.7 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 7.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.1 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 2.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.4 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.1 | 1.0 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 6.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.0 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 15.3 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 13.6 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.9 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 2.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 5.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.8 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.0 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.6 | GO:0031968 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.8 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.5 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 4.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.9 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 3.3 | GO:0019866 | mitochondrial inner membrane(GO:0005743) organelle inner membrane(GO:0019866) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.1 | 4.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 1.1 | 3.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.8 | 7.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.7 | 2.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.7 | 14.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.6 | 3.7 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.6 | 7.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.5 | 3.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.5 | 1.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.4 | 2.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.4 | 1.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.4 | 3.0 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.4 | 4.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 2.7 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.3 | 1.9 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.3 | 2.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 3.6 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.3 | 0.9 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.3 | 1.8 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.2 | 3.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.2 | 1.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.2 | 4.6 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 0.9 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 0.7 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 1.5 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 1.0 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 4.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.2 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.1 | 2.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 2.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 3.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.1 | 3.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 2.2 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 1.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.8 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 6.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.7 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 0.5 | GO:0005549 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.1 | 2.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.5 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.6 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 2.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 1.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 1.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.5 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 1.9 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.5 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 2.0 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 2.1 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.0 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 2.2 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.4 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.3 | GO:0046625 | sphingolipid binding(GO:0046625) ceramide binding(GO:0097001) |
| 0.0 | 0.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.6 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.7 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 1.9 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 6.0 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 2.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 4.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 5.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 10.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 14.4 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 2.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 4.6 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 1.1 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 3.9 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 8.1 | GO:0004175 | endopeptidase activity(GO:0004175) |
| 0.0 | 0.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 31.3 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 2.9 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 3.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 5.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 4.1 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 2.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 2.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 1.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 2.4 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.0 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.9 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.7 | PID P73PATHWAY | p73 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 6.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.5 | 2.6 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.5 | 7.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 4.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.2 | 5.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.2 | 7.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 2.5 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.2 | 6.8 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 2.0 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 2.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 3.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.9 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 1.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 3.3 | REACTOME ACTIVATION OF THE MRNA UPON BINDING OF THE CAP BINDING COMPLEX AND EIFS AND SUBSEQUENT BINDING TO 43S | Genes involved in Activation of the mRNA upon binding of the cap-binding complex and eIFs, and subsequent binding to 43S |
| 0.1 | 3.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 5.3 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 2.4 | REACTOME TRAF6 MEDIATED INDUCTION OF NFKB AND MAP KINASES UPON TLR7 8 OR 9 ACTIVATION | Genes involved in TRAF6 mediated induction of NFkB and MAP kinases upon TLR7/8 or 9 activation |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 1.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |