Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
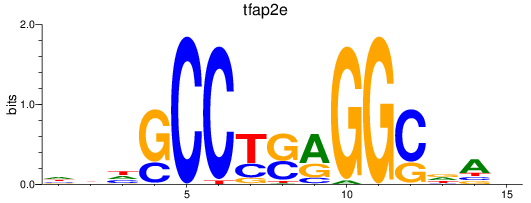
Results for tfap2e
Z-value: 0.44
Transcription factors associated with tfap2e
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2e
|
ENSDARG00000008861 | transcription factor AP-2 epsilon |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2e | dr11_v1_chr19_-_47456787_47456787 | 0.23 | 3.5e-01 | Click! |
Activity profile of tfap2e motif
Sorted Z-values of tfap2e motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_26704829 | 0.74 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr3_-_55328548 | 0.43 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr23_+_27778670 | 0.36 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr23_-_27647769 | 0.32 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr3_-_54607166 | 0.32 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr9_+_32358514 | 0.31 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr17_+_34186632 | 0.31 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr19_+_1184878 | 0.28 |
ENSDART00000163539
|
scrt1a
|
scratch family zinc finger 1a |
| chr7_-_23563092 | 0.28 |
ENSDART00000132275
|
gpr185b
|
G protein-coupled receptor 185 b |
| chr6_-_39313027 | 0.28 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr11_-_26180714 | 0.27 |
ENSDART00000173597
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr19_-_30510930 | 0.27 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr11_+_31323746 | 0.27 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr21_-_39058490 | 0.26 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr10_-_5135788 | 0.26 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr5_-_14509137 | 0.25 |
ENSDART00000180742
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr19_+_2835240 | 0.24 |
ENSDART00000190838
|
CDCP1
|
CUB domain containing protein 1 |
| chr19_-_31707892 | 0.23 |
ENSDART00000088427
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr10_+_5135842 | 0.23 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr24_+_42074143 | 0.23 |
ENSDART00000170514
|
top1mt
|
DNA topoisomerase I mitochondrial |
| chr15_+_44929497 | 0.22 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr14_-_33821515 | 0.21 |
ENSDART00000173218
|
vimr2
|
vimentin-related 2 |
| chr25_+_36405021 | 0.21 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr7_-_51476276 | 0.20 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr12_+_19036380 | 0.20 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr24_+_21346796 | 0.20 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr5_+_46424437 | 0.20 |
ENSDART00000186511
|
vcana
|
versican a |
| chr7_+_32693890 | 0.20 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr16_-_39859119 | 0.19 |
ENSDART00000138388
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr19_-_10395683 | 0.19 |
ENSDART00000109488
|
zgc:194578
|
zgc:194578 |
| chr6_-_10320676 | 0.19 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr13_+_16342926 | 0.18 |
ENSDART00000105496
ENSDART00000124710 |
dlg5a
|
discs, large homolog 5a (Drosophila) |
| chr25_+_5249513 | 0.18 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr16_-_46393154 | 0.17 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr1_+_33328857 | 0.17 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr8_-_11004726 | 0.16 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr16_-_5143124 | 0.16 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr14_+_39258569 | 0.16 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr17_-_15149192 | 0.15 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr4_+_32649955 | 0.15 |
ENSDART00000135750
|
znf1042
|
zinc finger protein 1042 |
| chr11_+_14333441 | 0.15 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr10_-_2526526 | 0.15 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr5_+_63302660 | 0.13 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr18_+_34478959 | 0.12 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr11_+_31324335 | 0.12 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr12_+_43585527 | 0.12 |
ENSDART00000161590
|
clrn3
|
clarin 3 |
| chr25_+_11281970 | 0.11 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr9_+_32978302 | 0.11 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr4_+_26972862 | 0.11 |
ENSDART00000011519
ENSDART00000138830 |
slc6a1l
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1, like |
| chr2_+_27010439 | 0.10 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr23_+_2760573 | 0.10 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr2_+_1988036 | 0.10 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr20_-_20312789 | 0.10 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr21_-_21089781 | 0.10 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr1_-_30457062 | 0.10 |
ENSDART00000185318
ENSDART00000157924 ENSDART00000161380 |
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr21_+_39941559 | 0.10 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr25_-_19574146 | 0.10 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr20_-_15089738 | 0.09 |
ENSDART00000164552
|
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr4_-_1914228 | 0.09 |
ENSDART00000087835
|
ano6
|
anoctamin 6 |
| chr4_+_77966055 | 0.09 |
ENSDART00000174203
ENSDART00000130100 ENSDART00000080665 ENSDART00000174317 ENSDART00000190123 |
zgc:113921
|
zgc:113921 |
| chr3_-_8160457 | 0.09 |
ENSDART00000183379
|
si:ch211-51i16.1
|
si:ch211-51i16.1 |
| chr23_-_18668836 | 0.09 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr7_-_15252540 | 0.09 |
ENSDART00000173072
ENSDART00000125258 |
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr18_-_47103131 | 0.09 |
ENSDART00000188553
|
CABZ01039304.1
|
|
| chr3_-_49163683 | 0.08 |
ENSDART00000166146
|
dnajb1a
|
DnaJ (Hsp40) homolog, subfamily B, member 1a |
| chr11_+_1657221 | 0.08 |
ENSDART00000164394
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr2_+_24199073 | 0.08 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr21_-_32680638 | 0.08 |
ENSDART00000147203
|
adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr20_-_2725930 | 0.06 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr16_-_29458806 | 0.06 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr23_-_9965148 | 0.06 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr2_+_17508323 | 0.05 |
ENSDART00000169703
|
pimr199
|
Pim proto-oncogene, serine/threonine kinase, related 199 |
| chr22_+_15633013 | 0.05 |
ENSDART00000188095
ENSDART00000048763 |
CABZ01090041.1
|
|
| chr14_-_31465905 | 0.05 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr18_+_34181655 | 0.05 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr11_-_28050559 | 0.05 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr19_+_32979331 | 0.05 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr24_-_6898302 | 0.04 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr6_-_25369295 | 0.04 |
ENSDART00000191475
|
PKN2 (1 of many)
|
zgc:153916 |
| chr5_+_27267186 | 0.04 |
ENSDART00000182238
ENSDART00000087857 |
unc5db
|
unc-5 netrin receptor Db |
| chr5_-_38451082 | 0.04 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr2_-_22927581 | 0.04 |
ENSDART00000109515
|
myo7bb
|
myosin VIIBb |
| chr1_-_53476758 | 0.03 |
ENSDART00000074221
|
b3gnt2a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2a |
| chr5_-_62801879 | 0.03 |
ENSDART00000191749
|
adora2b
|
adenosine A2b receptor |
| chr11_+_30253968 | 0.03 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr13_+_24279021 | 0.03 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr2_+_17556177 | 0.02 |
ENSDART00000074113
|
pimr197
|
Pim proto-oncogene, serine/threonine kinase, related 197 |
| chr24_-_21903588 | 0.02 |
ENSDART00000180991
|
spata13
|
spermatogenesis associated 13 |
| chr9_-_2892045 | 0.01 |
ENSDART00000137201
|
cdca7a
|
cell division cycle associated 7a |
| chr5_+_25736979 | 0.01 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr6_-_49873020 | 0.01 |
ENSDART00000148511
|
gnas
|
GNAS complex locus |
| chr5_-_62801724 | 0.00 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr2_+_36616830 | 0.00 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr24_-_21903360 | 0.00 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr22_+_110158 | 0.00 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr7_-_2116512 | 0.00 |
ENSDART00000098148
|
si:cabz01007802.1
|
si:cabz01007802.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2e
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0048909 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.1 | 0.2 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.2 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.2 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.4 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |