Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
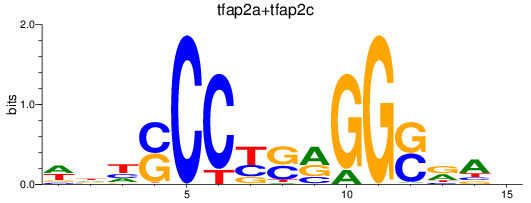
Results for tfap2a+tfap2c
Z-value: 0.61
Transcription factors associated with tfap2a+tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2c
|
ENSDARG00000040606 | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
|
tfap2a
|
ENSDARG00000059279 | transcription factor AP-2 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2c | dr11_v1_chr6_+_56147812_56147812 | 0.91 | 6.2e-08 | Click! |
| tfap2a | dr11_v1_chr24_-_8730913_8731039 | 0.63 | 4.0e-03 | Click! |
Activity profile of tfap2a+tfap2c motif
Sorted Z-values of tfap2a+tfap2c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_46424437 | 1.72 |
ENSDART00000186511
|
vcana
|
versican a |
| chr23_-_27152866 | 1.46 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr11_+_14333441 | 1.36 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr3_-_55328548 | 1.13 |
ENSDART00000082944
|
dock6
|
dedicator of cytokinesis 6 |
| chr24_-_21903588 | 1.04 |
ENSDART00000180991
|
spata13
|
spermatogenesis associated 13 |
| chr23_-_27647769 | 1.02 |
ENSDART00000059974
|
wnt10b
|
wingless-type MMTV integration site family, member 10b |
| chr3_+_26244353 | 1.01 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr24_-_21903360 | 1.00 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr7_+_32693890 | 0.97 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr3_-_54607166 | 0.95 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr11_+_31323746 | 0.92 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr11_+_14321113 | 0.90 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr20_-_2725594 | 0.90 |
ENSDART00000152120
|
akirin2
|
akirin 2 |
| chr16_-_5143124 | 0.87 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr25_+_5249513 | 0.86 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr5_+_63302660 | 0.83 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr20_-_2725930 | 0.82 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr3_-_26805455 | 0.81 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr24_-_37272116 | 0.81 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr11_+_31324335 | 0.78 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_+_14806851 | 0.77 |
ENSDART00000169235
|
fhdc2
|
FH2 domain containing 2 |
| chr8_+_48942470 | 0.73 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr23_-_9965148 | 0.71 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr2_+_27010439 | 0.71 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr8_+_48943009 | 0.65 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr7_-_34265481 | 0.64 |
ENSDART00000173596
|
si:ch211-98n17.5
|
si:ch211-98n17.5 |
| chr3_-_26244256 | 0.63 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr3_-_35865040 | 0.63 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr1_+_33328857 | 0.61 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr2_+_1988036 | 0.58 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr18_-_15551360 | 0.55 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr23_-_21215520 | 0.55 |
ENSDART00000143206
|
megf6a
|
multiple EGF-like-domains 6a |
| chr4_-_21466480 | 0.54 |
ENSDART00000139962
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr19_-_5369486 | 0.53 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr1_+_58990121 | 0.52 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr10_-_5135788 | 0.51 |
ENSDART00000108587
ENSDART00000138537 |
sec31a
|
SEC31 homolog A, COPII coat complex component |
| chr20_-_34292607 | 0.48 |
ENSDART00000144705
|
hmcn1
|
hemicentin 1 |
| chr10_-_8032885 | 0.47 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr5_-_62801879 | 0.43 |
ENSDART00000191749
|
adora2b
|
adenosine A2b receptor |
| chr22_+_30839702 | 0.43 |
ENSDART00000139849
ENSDART00000135775 |
si:dkey-49n23.3
|
si:dkey-49n23.3 |
| chr5_+_25736979 | 0.42 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr25_+_36405021 | 0.40 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr25_-_20378721 | 0.39 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr1_+_26467071 | 0.38 |
ENSDART00000112329
ENSDART00000159318 ENSDART00000193833 ENSDART00000011809 |
uso1
|
USO1 vesicle transport factor |
| chr18_+_34181655 | 0.38 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr1_+_27977297 | 0.38 |
ENSDART00000180692
ENSDART00000166819 |
sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr5_+_61799629 | 0.36 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr5_-_62801724 | 0.32 |
ENSDART00000144469
|
adora2b
|
adenosine A2b receptor |
| chr8_-_11004726 | 0.32 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr3_+_3454610 | 0.31 |
ENSDART00000024900
|
zgc:165453
|
zgc:165453 |
| chr20_-_15089738 | 0.31 |
ENSDART00000164552
|
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr4_-_16345227 | 0.28 |
ENSDART00000079521
|
kera
|
keratocan |
| chr20_-_34292285 | 0.28 |
ENSDART00000020389
|
hmcn1
|
hemicentin 1 |
| chr13_-_36566260 | 0.27 |
ENSDART00000030133
|
synj2bp
|
synaptojanin 2 binding protein |
| chr8_-_13211527 | 0.24 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr20_+_4157815 | 0.24 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr24_+_21346796 | 0.23 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr11_-_8782871 | 0.22 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr18_-_31969939 | 0.22 |
ENSDART00000171516
|
olfcs1
|
olfactory receptor C family, s1 |
| chr19_-_30510930 | 0.21 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr11_-_28050559 | 0.20 |
ENSDART00000136859
|
ece1
|
endothelin converting enzyme 1 |
| chr13_-_28674422 | 0.17 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr15_+_618081 | 0.17 |
ENSDART00000181518
|
si:ch211-210b2.1
|
si:ch211-210b2.1 |
| chr5_+_21100356 | 0.17 |
ENSDART00000051586
|
hs3st1l2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1-like 2 |
| chr7_-_15252540 | 0.16 |
ENSDART00000173072
ENSDART00000125258 |
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr16_-_29458806 | 0.15 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr2_-_43635777 | 0.13 |
ENSDART00000148633
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr15_+_44929497 | 0.13 |
ENSDART00000111373
|
pdgfd
|
platelet derived growth factor d |
| chr8_+_48858132 | 0.13 |
ENSDART00000124737
ENSDART00000079644 |
tp73
|
tumor protein p73 |
| chr24_+_31374324 | 0.13 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr3_+_36160979 | 0.12 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr9_-_98982 | 0.12 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr5_-_38451082 | 0.12 |
ENSDART00000136428
|
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr4_-_68853063 | 0.10 |
ENSDART00000162386
|
si:dkey-264f17.1
|
si:dkey-264f17.1 |
| chr15_+_6459847 | 0.10 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr14_+_14806692 | 0.10 |
ENSDART00000193050
|
fhdc2
|
FH2 domain containing 2 |
| chr8_-_4789034 | 0.08 |
ENSDART00000082256
|
ciao1
|
cytosolic iron-sulfur assembly component 1 |
| chr16_+_26601364 | 0.07 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr1_+_16548733 | 0.04 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr8_-_26792912 | 0.04 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr16_-_46393154 | 0.03 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr5_+_6672870 | 0.02 |
ENSDART00000126598
|
pxna
|
paxillin a |
| chr11_-_26180714 | 0.01 |
ENSDART00000173597
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr20_-_20312789 | 0.00 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2a+tfap2c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0003093 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.3 | 0.9 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 0.8 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.2 | 1.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 1.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 0.2 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.7 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.4 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.5 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 1.7 | GO:0045089 | positive regulation of innate immune response(GO:0045089) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.8 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 2.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0097361 | CIA complex(GO:0097361) |
| 0.0 | 1.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.5 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 3.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.0 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |