Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
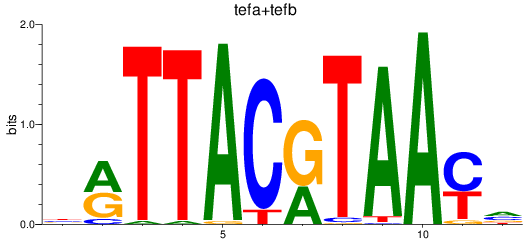
Results for tefa+tefb
Z-value: 1.39
Transcription factors associated with tefa+tefb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tefa
|
ENSDARG00000039117 | TEF transcription factor, PAR bZIP family member a |
|
tefb
|
ENSDARG00000098103 | TEF transcription factor, PAR bZIP family member b |
|
tefa
|
ENSDARG00000109532 | TEF transcription factor, PAR bZIP family member a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tefb | dr11_v1_chr3_-_5067585_5067585 | 0.91 | 9.7e-08 | Click! |
| tefa | dr11_v1_chr12_-_19151708_19151708 | -0.39 | 1.0e-01 | Click! |
Activity profile of tefa+tefb motif
Sorted Z-values of tefa+tefb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_44953349 | 5.40 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr9_-_48736388 | 5.28 |
ENSDART00000022074
|
dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr25_+_15354095 | 5.15 |
ENSDART00000090397
|
kiaa1549la
|
KIAA1549-like a |
| chr20_-_1151265 | 4.32 |
ENSDART00000012376
|
gabrr1
|
gamma-aminobutyric acid (GABA) A receptor, rho 1 |
| chr2_+_24177190 | 4.15 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr20_+_1121458 | 4.07 |
ENSDART00000064472
|
pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chr20_-_25533739 | 4.01 |
ENSDART00000063064
|
cyp2ad6
|
cytochrome P450, family 2, subfamily AD, polypeptide 6 |
| chr24_-_39772045 | 3.85 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr11_-_10770053 | 3.74 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr9_-_44953664 | 3.59 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr12_-_26383242 | 3.56 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr3_+_7763114 | 3.48 |
ENSDART00000057434
|
hook2
|
hook microtubule-tethering protein 2 |
| chr7_-_6604623 | 3.25 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr11_+_6819050 | 3.22 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr21_+_43178831 | 3.17 |
ENSDART00000151512
|
aff4
|
AF4/FMR2 family, member 4 |
| chr20_-_45661049 | 3.17 |
ENSDART00000124582
ENSDART00000131251 |
napbb
|
N-ethylmaleimide-sensitive factor attachment protein, beta b |
| chr23_+_4022620 | 3.11 |
ENSDART00000055099
|
b4galt5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr7_+_17716601 | 3.06 |
ENSDART00000173792
ENSDART00000080825 |
rtn3
|
reticulon 3 |
| chr20_-_44576949 | 2.97 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr20_+_23083800 | 2.86 |
ENSDART00000132093
|
usp46
|
ubiquitin specific peptidase 46 |
| chr9_+_13682133 | 2.85 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr3_-_28665291 | 2.83 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr5_+_1278092 | 2.83 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr16_+_46111849 | 2.82 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr9_+_46644633 | 2.78 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr7_+_38717624 | 2.78 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr20_+_38201644 | 2.77 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr7_+_4162994 | 2.77 |
ENSDART00000172800
|
si:ch211-63p21.1
|
si:ch211-63p21.1 |
| chr9_-_16109001 | 2.71 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr17_+_15845765 | 2.69 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr6_-_1553314 | 2.52 |
ENSDART00000077209
|
tpra1
|
transmembrane protein, adipocyte asscociated 1 |
| chr16_+_39146696 | 2.48 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr5_-_22001003 | 2.41 |
ENSDART00000134393
ENSDART00000143878 |
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr7_-_32833153 | 2.40 |
ENSDART00000099871
ENSDART00000099872 |
slc17a6b
|
solute carrier family 17 (vesicular glutamate transporter), member 6b |
| chr16_-_45069882 | 2.40 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr10_-_24371312 | 2.39 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr14_+_4151379 | 2.34 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr5_-_21970881 | 2.21 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr23_+_33963619 | 2.13 |
ENSDART00000140666
ENSDART00000084792 |
plpbp
|
pyridoxal phosphate binding protein |
| chr2_+_7192966 | 2.10 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr5_+_69808763 | 1.95 |
ENSDART00000143482
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr19_+_4990320 | 1.95 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr21_-_42202792 | 1.90 |
ENSDART00000124708
|
gabra6b
|
gamma-aminobutyric acid (GABA) A receptor, alpha 6b |
| chr15_-_24178893 | 1.86 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr1_+_54673846 | 1.84 |
ENSDART00000145018
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr11_-_23459779 | 1.82 |
ENSDART00000183935
ENSDART00000184125 ENSDART00000193284 |
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr23_-_21763598 | 1.78 |
ENSDART00000145408
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr18_+_25546227 | 1.74 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr6_-_29612269 | 1.73 |
ENSDART00000104293
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr23_+_19590006 | 1.73 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr5_+_15350954 | 1.71 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr19_+_4990496 | 1.71 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr17_-_23709347 | 1.65 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr5_-_21030934 | 1.64 |
ENSDART00000133461
ENSDART00000098667 |
camk2b1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 1 |
| chr19_-_45650994 | 1.63 |
ENSDART00000163504
|
trps1
|
trichorhinophalangeal syndrome I |
| chr7_-_40959667 | 1.61 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr11_-_7078392 | 1.56 |
ENSDART00000112156
ENSDART00000188556 |
si:ch211-253b8.5
|
si:ch211-253b8.5 |
| chr18_-_2549198 | 1.54 |
ENSDART00000186516
|
CABZ01070631.1
|
|
| chr21_-_20733615 | 1.50 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr16_-_35427060 | 1.46 |
ENSDART00000172294
|
ctps1b
|
CTP synthase 1b |
| chr12_+_15002757 | 1.41 |
ENSDART00000135036
|
mylpfb
|
myosin light chain, phosphorylatable, fast skeletal muscle b |
| chr17_-_43466317 | 1.41 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr6_-_39649504 | 1.37 |
ENSDART00000179960
ENSDART00000190951 |
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr17_-_20979077 | 1.33 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr25_+_34581494 | 1.32 |
ENSDART00000167690
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr8_+_8712446 | 1.31 |
ENSDART00000158674
|
elk1
|
ELK1, member of ETS oncogene family |
| chr1_+_31112436 | 1.31 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr14_+_36628131 | 1.26 |
ENSDART00000188625
ENSDART00000125345 |
TENM3
|
si:dkey-237h12.3 |
| chr10_+_33895315 | 1.20 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr7_+_34297271 | 1.20 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr5_-_29238889 | 1.19 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr7_+_38278860 | 1.16 |
ENSDART00000016265
|
lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr15_+_28303161 | 1.14 |
ENSDART00000087926
|
myo1cb
|
myosin Ic, paralog b |
| chr4_-_5302866 | 1.11 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr5_-_30074332 | 1.09 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr4_-_14915268 | 1.07 |
ENSDART00000067040
|
si:dkey-180p18.9
|
si:dkey-180p18.9 |
| chr4_-_5302162 | 1.06 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr19_+_7735157 | 1.05 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr8_-_29851706 | 1.03 |
ENSDART00000149297
|
slc20a2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr22_-_13165186 | 1.02 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr9_-_5318873 | 0.98 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr18_-_1228688 | 0.98 |
ENSDART00000064403
|
nptnb
|
neuroplastin b |
| chr22_-_18491813 | 0.97 |
ENSDART00000105419
|
si:ch211-212d10.2
|
si:ch211-212d10.2 |
| chr16_-_19568388 | 0.95 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr15_+_37559570 | 0.92 |
ENSDART00000085522
|
hspb6
|
heat shock protein, alpha-crystallin-related, b6 |
| chr17_+_49281597 | 0.88 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr15_-_16121496 | 0.87 |
ENSDART00000128624
|
sgk494a
|
uncharacterized serine/threonine-protein kinase SgK494a |
| chr5_-_69212184 | 0.81 |
ENSDART00000053963
|
mat2ab
|
methionine adenosyltransferase II, alpha b |
| chr11_+_25504215 | 0.76 |
ENSDART00000154213
|
tfe3b
|
transcription factor binding to IGHM enhancer 3b |
| chr1_+_19764995 | 0.76 |
ENSDART00000138276
|
si:ch211-42i9.8
|
si:ch211-42i9.8 |
| chr5_-_65153731 | 0.65 |
ENSDART00000171024
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr17_-_31308658 | 0.64 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr25_-_11088839 | 0.64 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr14_+_21699414 | 0.61 |
ENSDART00000169942
|
stx3a
|
syntaxin 3A |
| chr9_-_13871935 | 0.59 |
ENSDART00000146597
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr1_+_16600690 | 0.57 |
ENSDART00000162164
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr1_-_354115 | 0.56 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr18_-_39288894 | 0.50 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr23_+_9522781 | 0.47 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr7_+_30392613 | 0.44 |
ENSDART00000075508
|
lipca
|
lipase, hepatic a |
| chr16_-_54800202 | 0.44 |
ENSDART00000059560
ENSDART00000161833 |
khdc4
|
KH domain containing 4, pre-mRNA splicing factor |
| chr25_-_7887274 | 0.43 |
ENSDART00000157276
|
si:ch73-151m17.5
|
si:ch73-151m17.5 |
| chr25_-_16826219 | 0.41 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr12_-_23365737 | 0.41 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr16_+_34523515 | 0.41 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr11_-_40147032 | 0.36 |
ENSDART00000052918
|
si:dkey-264d12.4
|
si:dkey-264d12.4 |
| chr21_-_39931285 | 0.35 |
ENSDART00000180010
ENSDART00000024407 |
tmigd1
|
transmembrane and immunoglobulin domain containing 1 |
| chr25_-_12803723 | 0.25 |
ENSDART00000158787
|
ca5a
|
carbonic anhydrase Va |
| chr21_+_21374277 | 0.22 |
ENSDART00000079431
|
rtn2b
|
reticulon 2b |
| chr7_-_38792543 | 0.21 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr6_+_50451337 | 0.17 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr20_-_44496245 | 0.14 |
ENSDART00000012229
|
fkbp1b
|
FK506 binding protein 1b |
| chr22_-_37686966 | 0.11 |
ENSDART00000192217
|
htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
Network of associatons between targets according to the STRING database.
First level regulatory network of tefa+tefb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 9.0 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 1.1 | 4.3 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 1.1 | 3.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.8 | 3.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.6 | 2.5 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.5 | 1.6 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.5 | 3.3 | GO:0036268 | swimming(GO:0036268) |
| 0.5 | 1.9 | GO:0046440 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.4 | 8.0 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.4 | 1.5 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.3 | 2.9 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.3 | 1.0 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.3 | 2.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.3 | 1.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.3 | 6.5 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.3 | 2.7 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.3 | 1.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 3.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 3.7 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.2 | 0.6 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.2 | 3.0 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.2 | 1.7 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.2 | 1.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 2.8 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.9 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.8 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.0 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.1 | 1.0 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 4.0 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 2.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 2.8 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 0.8 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 3.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.0 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 2.7 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.5 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 2.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 2.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 2.4 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 2.1 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.3 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 3.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 1.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.3 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.7 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 4.2 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.0 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 3.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 3.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.4 | 1.5 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.3 | 1.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.3 | 3.7 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.3 | 8.9 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 1.7 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.2 | 0.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.2 | 2.4 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.8 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 2.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 2.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 3.5 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 5.8 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 4.6 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.1 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.0 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 3.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 7.7 | GO:0043005 | neuron projection(GO:0043005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.7 | 5.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.6 | 2.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 1.6 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.5 | 3.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.4 | 1.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.4 | 1.5 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.3 | 2.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.3 | 1.9 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.3 | 6.5 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.3 | 8.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.2 | 1.7 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.8 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.2 | 2.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 9.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.4 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.0 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 2.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 4.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 5.5 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 3.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.8 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 2.7 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 1.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 11.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.5 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 2.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.0 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 4.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.7 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 4.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 3.1 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 2.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.8 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |