Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
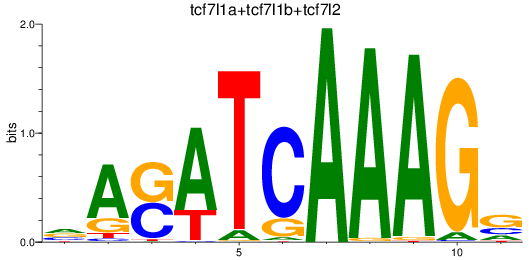
Results for tcf7l1a+tcf7l1b+tcf7l2
Z-value: 1.37
Transcription factors associated with tcf7l1a+tcf7l1b+tcf7l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7l2
|
ENSDARG00000004415 | transcription factor 7 like 2 |
|
tcf7l1b
|
ENSDARG00000007369 | transcription factor 7 like 1b |
|
tcf7l1a
|
ENSDARG00000038159 | transcription factor 7 like 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7l2 | dr11_v1_chr12_-_31103187_31103316 | 0.89 | 3.2e-07 | Click! |
| tcf7l1a | dr11_v1_chr10_-_42297889_42297889 | -0.86 | 2.4e-06 | Click! |
| tcf7l1b | dr11_v1_chr8_-_52143109_52143109 | -0.51 | 2.7e-02 | Click! |
Activity profile of tcf7l1a+tcf7l1b+tcf7l2 motif
Sorted Z-values of tcf7l1a+tcf7l1b+tcf7l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_6109861 | 6.69 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr16_+_52512025 | 5.46 |
ENSDART00000056095
|
fabp10a
|
fatty acid binding protein 10a, liver basic |
| chr7_+_39446247 | 5.14 |
ENSDART00000033610
ENSDART00000099015 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr13_+_3252950 | 4.70 |
ENSDART00000020671
|
prph2b
|
peripherin 2b (retinal degeneration, slow) |
| chr12_+_5129245 | 4.50 |
ENSDART00000169073
|
pde6c
|
phosphodiesterase 6C, cGMP-specific, cone, alpha prime |
| chr17_-_20897407 | 4.19 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr25_-_30429607 | 4.07 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr22_-_282498 | 3.93 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr20_+_35484070 | 3.92 |
ENSDART00000026234
ENSDART00000141675 |
mep1a.2
|
meprin A, alpha (PABA peptide hydrolase), tandem duplicate 2 |
| chr25_+_14092871 | 3.81 |
ENSDART00000067239
|
guca1g
|
guanylate cyclase activator 1g |
| chr22_-_263117 | 3.67 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr17_+_27176243 | 3.66 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr15_-_12545683 | 3.65 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr11_+_41135055 | 3.58 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr13_+_27073901 | 3.53 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr5_+_37068223 | 3.43 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr15_+_30158652 | 3.41 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr14_+_22172047 | 3.40 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr23_-_39784368 | 3.23 |
ENSDART00000110282
|
si:ch211-286f9.2
|
si:ch211-286f9.2 |
| chr12_+_40905427 | 3.22 |
ENSDART00000170526
ENSDART00000185771 ENSDART00000193945 |
CDH18
|
cadherin 18 |
| chr8_-_25329967 | 3.10 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr16_-_383664 | 3.06 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr9_-_9992697 | 3.04 |
ENSDART00000123415
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr20_+_30445971 | 2.99 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr21_-_37889727 | 2.97 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr20_+_43083745 | 2.90 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr3_-_30861177 | 2.82 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr22_-_237651 | 2.82 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr22_-_294700 | 2.81 |
ENSDART00000189179
|
CABZ01079179.1
|
|
| chr25_+_7229046 | 2.80 |
ENSDART00000149965
ENSDART00000041820 |
lingo1a
|
leucine rich repeat and Ig domain containing 1a |
| chr9_-_9998087 | 2.79 |
ENSDART00000124423
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr5_-_18474486 | 2.76 |
ENSDART00000090580
|
si:dkey-215k6.1
|
si:dkey-215k6.1 |
| chr5_+_15992655 | 2.71 |
ENSDART00000182148
|
znrf3
|
zinc and ring finger 3 |
| chr20_-_40451115 | 2.56 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr12_-_31103187 | 2.56 |
ENSDART00000005562
ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 ENSDART00000122972 ENSDART00000153068 |
tcf7l2
|
transcription factor 7 like 2 |
| chr15_-_2188332 | 2.52 |
ENSDART00000138941
ENSDART00000009564 |
shox2
|
short stature homeobox 2 |
| chr24_-_5932982 | 2.52 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr17_-_20897250 | 2.52 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr7_+_27977065 | 2.40 |
ENSDART00000089574
|
tub
|
tubby bipartite transcription factor |
| chr2_-_30784502 | 2.39 |
ENSDART00000056735
|
rgs20
|
regulator of G protein signaling 20 |
| chr13_-_23264724 | 2.37 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr20_+_34455645 | 2.34 |
ENSDART00000135789
|
mettl11b
|
methyltransferase like 11B |
| chr8_-_18899427 | 2.21 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr13_+_28702104 | 2.18 |
ENSDART00000135481
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr18_+_10907999 | 2.16 |
ENSDART00000133555
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr6_-_57938043 | 2.16 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr12_+_35203091 | 2.14 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr3_-_33573252 | 2.10 |
ENSDART00000191339
ENSDART00000184612 ENSDART00000187232 ENSDART00000190671 |
CR847537.1
|
|
| chr2_+_9822319 | 2.10 |
ENSDART00000144078
ENSDART00000144371 |
anxa13l
|
annexin A13, like |
| chr7_-_30087048 | 2.08 |
ENSDART00000112743
|
nmbb
|
neuromedin Bb |
| chr8_+_17009199 | 2.07 |
ENSDART00000139452
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr25_-_19395476 | 2.06 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr10_-_39011514 | 2.06 |
ENSDART00000075123
|
pcp4a
|
Purkinje cell protein 4a |
| chr19_-_44801918 | 2.06 |
ENSDART00000108507
|
CSMD3 (1 of many)
|
si:ch211-233f16.1 |
| chr23_+_16620801 | 2.05 |
ENSDART00000189859
ENSDART00000184578 |
snphb
|
syntaphilin b |
| chr17_-_8638713 | 2.05 |
ENSDART00000148971
|
ctbp2a
|
C-terminal binding protein 2a |
| chr12_+_6214041 | 2.02 |
ENSDART00000179759
|
prkg1b
|
protein kinase, cGMP-dependent, type Ib |
| chr1_-_29045426 | 2.01 |
ENSDART00000019770
|
gpm6ba
|
glycoprotein M6Ba |
| chr6_-_15604157 | 1.96 |
ENSDART00000141597
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr24_+_32411753 | 1.95 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr4_+_10366532 | 1.87 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr23_-_20764227 | 1.86 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr7_+_34492744 | 1.85 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr2_+_2503396 | 1.81 |
ENSDART00000168418
|
crhr2
|
corticotropin releasing hormone receptor 2 |
| chr20_+_43942278 | 1.81 |
ENSDART00000100571
|
clic5b
|
chloride intracellular channel 5b |
| chr24_-_1985007 | 1.79 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr2_+_7192966 | 1.79 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr12_-_6880694 | 1.78 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr2_-_34483597 | 1.77 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr15_-_43768776 | 1.77 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr4_-_17409533 | 1.77 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr25_+_26921480 | 1.77 |
ENSDART00000155949
|
grm8b
|
glutamate receptor, metabotropic 8b |
| chr17_-_26507289 | 1.75 |
ENSDART00000155616
|
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr1_-_19233890 | 1.75 |
ENSDART00000127145
|
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr7_-_71486162 | 1.75 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr2_-_43083749 | 1.75 |
ENSDART00000084303
ENSDART00000145494 |
kcnq3
|
potassium voltage-gated channel, KQT-like subfamily, member 3 |
| chr20_+_41549200 | 1.72 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr15_-_163586 | 1.71 |
ENSDART00000163597
|
SEPT4
|
septin-4 |
| chr24_+_15897717 | 1.69 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr1_+_23161369 | 1.69 |
ENSDART00000162827
|
si:dkey-92j12.5
|
si:dkey-92j12.5 |
| chr8_+_14778292 | 1.68 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr4_+_7391110 | 1.68 |
ENSDART00000160708
ENSDART00000187823 |
tnni4a
|
troponin I4a |
| chr6_-_15603675 | 1.68 |
ENSDART00000143502
|
lrrfip1b
|
leucine rich repeat (in FLII) interacting protein 1b |
| chr2_-_30784198 | 1.68 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr13_+_28701233 | 1.66 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr12_-_33706726 | 1.65 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr24_-_32408404 | 1.65 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr5_+_26728115 | 1.63 |
ENSDART00000098545
|
tmem150aa
|
transmembrane protein 150Aa |
| chr1_+_32528097 | 1.63 |
ENSDART00000128317
|
nlgn4a
|
neuroligin 4a |
| chr2_-_30734098 | 1.62 |
ENSDART00000133769
|
rp1
|
retinitis pigmentosa 1 (autosomal dominant) |
| chr24_+_11105786 | 1.59 |
ENSDART00000175182
|
prlh2
|
prolactin releasing hormone 2 |
| chr2_+_30379650 | 1.59 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr19_+_42469058 | 1.58 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr9_+_28693592 | 1.54 |
ENSDART00000110198
|
zgc:162780
|
zgc:162780 |
| chr7_+_27976448 | 1.53 |
ENSDART00000181026
|
tub
|
tubby bipartite transcription factor |
| chr6_-_40429411 | 1.52 |
ENSDART00000156005
ENSDART00000156357 |
si:dkey-28n18.9
|
si:dkey-28n18.9 |
| chr1_-_8428736 | 1.52 |
ENSDART00000138435
ENSDART00000121823 |
syngr3b
|
synaptogyrin 3b |
| chr13_-_18835254 | 1.48 |
ENSDART00000147579
ENSDART00000146795 |
lzts2a
|
leucine zipper, putative tumor suppressor 2a |
| chr22_-_21150845 | 1.45 |
ENSDART00000027345
|
tmem59l
|
transmembrane protein 59-like |
| chr9_-_35155089 | 1.45 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr17_+_26611929 | 1.44 |
ENSDART00000166450
ENSDART00000087023 |
ttc7b
|
tetratricopeptide repeat domain 7B |
| chr8_-_22739757 | 1.44 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr4_+_69619 | 1.43 |
ENSDART00000164425
|
mansc1
|
MANSC domain containing 1 |
| chr12_-_31457301 | 1.41 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr16_+_17389116 | 1.41 |
ENSDART00000103750
ENSDART00000173448 |
fam131bb
|
family with sequence similarity 131, member Bb |
| chr21_-_40782393 | 1.40 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr23_+_37579107 | 1.40 |
ENSDART00000169376
|
plekhg5b
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5b |
| chr16_+_16933002 | 1.39 |
ENSDART00000173163
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr21_-_25522906 | 1.38 |
ENSDART00000110923
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr6_-_53143667 | 1.37 |
ENSDART00000079694
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr19_-_8732037 | 1.34 |
ENSDART00000138971
|
si:ch211-39a7.1
|
si:ch211-39a7.1 |
| chr8_-_34051548 | 1.32 |
ENSDART00000105204
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr15_-_30714130 | 1.31 |
ENSDART00000156914
ENSDART00000154714 |
msi2b
|
musashi RNA-binding protein 2b |
| chr12_-_31995840 | 1.31 |
ENSDART00000112881
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr19_-_6134802 | 1.31 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr12_+_10115964 | 1.30 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr7_+_19762595 | 1.30 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr20_+_25597527 | 1.25 |
ENSDART00000130242
|
cyp2j20
|
cytochrome P450, family 2, subfamily J, polypeptide 20 |
| chr16_+_39146696 | 1.24 |
ENSDART00000121756
ENSDART00000084381 |
sybu
|
syntabulin (syntaxin-interacting) |
| chr11_+_34522554 | 1.23 |
ENSDART00000109833
|
zmat3
|
zinc finger, matrin-type 3 |
| chr21_+_26612777 | 1.21 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr18_-_46380471 | 1.20 |
ENSDART00000144444
ENSDART00000086772 |
slc19a3b
|
solute carrier family 19 (thiamine transporter), member 3b |
| chr24_-_6546479 | 1.19 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr20_+_36234335 | 1.19 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr13_-_31829786 | 1.18 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr20_-_9436521 | 1.17 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr6_+_3282809 | 1.17 |
ENSDART00000187444
ENSDART00000187407 ENSDART00000191883 |
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr21_-_25722834 | 1.16 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr3_-_19091024 | 1.16 |
ENSDART00000188485
ENSDART00000110554 |
grin2ca
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Ca |
| chr18_+_45796096 | 1.16 |
ENSDART00000087070
|
abcc5
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 5 |
| chr15_-_31588162 | 1.15 |
ENSDART00000153598
|
hsph1
|
heat shock 105/110 protein 1 |
| chr16_+_39159752 | 1.14 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr22_+_3045495 | 1.10 |
ENSDART00000164061
|
LO017843.1
|
|
| chr7_-_42206720 | 1.10 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr9_+_10014817 | 1.09 |
ENSDART00000132065
|
nxph2a
|
neurexophilin 2a |
| chr13_+_4505232 | 1.08 |
ENSDART00000007500
ENSDART00000161684 |
pde10a
|
phosphodiesterase 10A |
| chr7_-_9674073 | 1.07 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr4_-_77432218 | 1.06 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr6_-_7208119 | 1.06 |
ENSDART00000105148
ENSDART00000185846 |
hs6st3a
|
heparan sulfate 6-O-sulfotransferase 3a |
| chr3_+_15773991 | 1.05 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr18_+_36806545 | 1.05 |
ENSDART00000098958
|
ttc9b
|
tetratricopeptide repeat domain 9B |
| chr8_+_36803415 | 1.03 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr6_-_26559921 | 1.03 |
ENSDART00000104532
|
sox14
|
SRY (sex determining region Y)-box 14 |
| chr20_+_39283849 | 1.01 |
ENSDART00000002481
ENSDART00000146683 |
scara3
|
scavenger receptor class A, member 3 |
| chr1_-_42779075 | 1.00 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr20_+_28266892 | 0.99 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr24_-_24848612 | 0.96 |
ENSDART00000190941
|
crhb
|
corticotropin releasing hormone b |
| chr3_+_55288200 | 0.96 |
ENSDART00000157002
ENSDART00000155600 |
si:dkey-114l24.2
|
si:dkey-114l24.2 |
| chr8_-_4097722 | 0.95 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr9_+_10014514 | 0.95 |
ENSDART00000185590
|
nxph2a
|
neurexophilin 2a |
| chr19_-_32600138 | 0.95 |
ENSDART00000052101
|
zgc:91944
|
zgc:91944 |
| chr12_+_32323098 | 0.94 |
ENSDART00000188722
|
ANKFN1
|
si:ch211-277e21.2 |
| chr8_-_50346246 | 0.93 |
ENSDART00000025008
|
stc1
|
stanniocalcin 1 |
| chr6_-_53144336 | 0.92 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr2_-_42396592 | 0.92 |
ENSDART00000127136
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr4_-_6459863 | 0.88 |
ENSDART00000138367
|
foxp2
|
forkhead box P2 |
| chr20_+_22666548 | 0.88 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr18_+_16330720 | 0.87 |
ENSDART00000080638
|
nts
|
neurotensin |
| chr25_-_19395156 | 0.86 |
ENSDART00000155335
|
map1ab
|
microtubule-associated protein 1Ab |
| chr14_+_30910114 | 0.86 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr5_+_44190974 | 0.84 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr21_-_28439596 | 0.84 |
ENSDART00000089980
ENSDART00000132844 |
rasgrp2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr13_+_4505079 | 0.83 |
ENSDART00000144312
|
pde10a
|
phosphodiesterase 10A |
| chr10_+_11309814 | 0.83 |
ENSDART00000145673
|
si:ch211-126i22.5
|
si:ch211-126i22.5 |
| chr15_+_24691088 | 0.82 |
ENSDART00000110618
|
LRRC75A
|
si:dkey-151p21.7 |
| chr16_+_34493987 | 0.81 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr7_+_42206847 | 0.80 |
ENSDART00000149250
|
phkb
|
phosphorylase kinase, beta |
| chr6_+_58915889 | 0.80 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr15_+_28106498 | 0.78 |
ENSDART00000041707
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr16_+_19014886 | 0.76 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr19_-_10043142 | 0.75 |
ENSDART00000193016
|
grin2da
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2D, a |
| chr9_-_14055959 | 0.74 |
ENSDART00000146675
|
fer1l6
|
fer-1-like family member 6 |
| chr3_-_41715690 | 0.73 |
ENSDART00000184703
|
BX324110.1
|
|
| chr15_+_22722684 | 0.73 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr3_-_41292275 | 0.71 |
ENSDART00000144088
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr15_+_1705167 | 0.70 |
ENSDART00000081940
|
otol1b
|
otolin 1b |
| chr13_-_24906307 | 0.70 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr5_-_8096232 | 0.69 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr9_+_3055566 | 0.69 |
ENSDART00000189906
ENSDART00000175891 ENSDART00000093021 |
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr7_+_56651759 | 0.69 |
ENSDART00000073600
|
kcng4b
|
potassium voltage-gated channel, subfamily G, member 4b |
| chr13_-_2112450 | 0.68 |
ENSDART00000189343
|
fam83b
|
family with sequence similarity 83, member B |
| chr17_+_3379673 | 0.67 |
ENSDART00000176354
|
sntg2
|
syntrophin, gamma 2 |
| chr4_-_2036620 | 0.66 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr13_-_31441042 | 0.66 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr11_+_34523132 | 0.66 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr8_-_34052019 | 0.63 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr9_+_3087889 | 0.63 |
ENSDART00000146148
|
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr5_-_27867657 | 0.63 |
ENSDART00000112495
|
tcima
|
transcriptional and immune response regulator a |
| chr4_+_9836465 | 0.61 |
ENSDART00000004879
|
hsp90b1
|
heat shock protein 90, beta (grp94), member 1 |
| chr19_-_32487469 | 0.61 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr6_-_13498745 | 0.61 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr5_-_28149767 | 0.61 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr1_-_26023678 | 0.60 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr17_-_24714837 | 0.60 |
ENSDART00000154871
|
si:ch211-15d5.11
|
si:ch211-15d5.11 |
| chr21_-_25522510 | 0.59 |
ENSDART00000162711
|
cnksr2b
|
connector enhancer of kinase suppressor of Ras 2b |
| chr3_-_41292569 | 0.59 |
ENSDART00000111856
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr23_-_29824146 | 0.58 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr9_-_12034444 | 0.57 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr11_-_18604317 | 0.55 |
ENSDART00000182081
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr20_+_36730049 | 0.55 |
ENSDART00000045948
|
ncoa1
|
nuclear receptor coactivator 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7l1a+tcf7l1b+tcf7l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.7 | 0.7 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.7 | 2.7 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.7 | 2.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.6 | 2.6 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.6 | 2.4 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.6 | 2.4 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.6 | 1.8 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.5 | 3.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.5 | 2.9 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.5 | 1.4 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.4 | 1.7 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.4 | 1.3 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.4 | 2.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.4 | 2.1 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.4 | 1.9 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.3 | 5.0 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.3 | 1.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.3 | 2.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.2 | 2.3 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.2 | 5.1 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 2.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.2 | 1.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 0.5 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.2 | 1.8 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 1.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.2 | 0.7 | GO:0090579 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 3.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 1.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 4.5 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 2.1 | GO:0046887 | positive regulation of hormone secretion(GO:0046887) |
| 0.1 | 0.5 | GO:0060092 | inhibitory postsynaptic potential(GO:0060080) regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.7 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 1.2 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 2.9 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.8 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 2.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.6 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 0.8 | GO:1900186 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 1.0 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.5 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 1.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 3.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.4 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.5 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.3 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.1 | 1.7 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 3.4 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 2.8 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.1 | 3.9 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 4.8 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.1 | 1.8 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 1.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 2.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.0 | 0.3 | GO:0015846 | polyamine transport(GO:0015846) putrescine biosynthetic process from ornithine(GO:0033387) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.5 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 1.6 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.6 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 1.4 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.3 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 1.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 1.9 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.9 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 2.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.5 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.4 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 2.5 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.1 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 5.2 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 5.2 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.2 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 4.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 1.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.7 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 2.1 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 1.1 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.2 | GO:0048512 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.2 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.6 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.2 | 0.7 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 2.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 6.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 3.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.7 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 1.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 3.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.8 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 2.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 8.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.6 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.8 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 1.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 1.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 6.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 2.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.7 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.7 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.2 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 3.9 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.3 | GO:0014069 | postsynaptic density(GO:0014069) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.5 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.8 | 2.3 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.6 | 1.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.5 | 2.9 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.5 | 3.8 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.5 | 2.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.5 | 5.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 1.3 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.4 | 3.5 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 1.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.3 | 2.0 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.3 | 1.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.3 | 2.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.3 | 2.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.3 | 1.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.3 | 1.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.3 | 1.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.2 | 1.0 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.2 | 0.6 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.2 | 1.8 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.2 | 1.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.7 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.2 | 1.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 6.6 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.2 | 3.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 3.6 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.5 | GO:0015105 | arsenite transmembrane transporter activity(GO:0015105) |
| 0.1 | 4.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 5.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.2 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.1 | 1.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 3.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 3.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 0.9 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.8 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 1.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.5 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.2 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 1.8 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.5 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 2.2 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 7.5 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.4 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 4.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.7 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 3.6 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 1.0 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 1.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 1.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 2.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.5 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.1 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.2 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 3.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 1.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 3.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 4.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 0.8 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.2 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 1.9 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 0.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.9 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |