Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
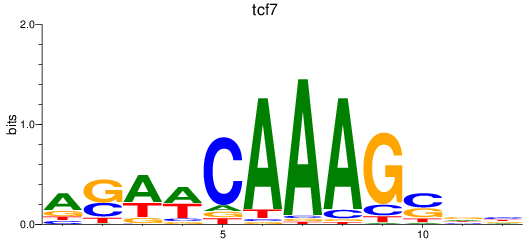
Results for tcf7
Z-value: 0.49
Transcription factors associated with tcf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf7
|
ENSDARG00000038672 | transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf7 | dr11_v1_chr21_+_45268112_45268112 | -0.48 | 3.8e-02 | Click! |
Activity profile of tcf7 motif
Sorted Z-values of tcf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_35075847 | 3.22 |
ENSDART00000193469
ENSDART00000037346 |
ctrb1
|
chymotrypsinogen B1 |
| chr19_+_40856534 | 2.00 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr20_+_46040666 | 1.98 |
ENSDART00000060744
|
si:dkey-7c18.24
|
si:dkey-7c18.24 |
| chr9_-_22232902 | 1.85 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr6_-_35487413 | 1.70 |
ENSDART00000102461
|
rgs8
|
regulator of G protein signaling 8 |
| chr15_+_6109861 | 1.50 |
ENSDART00000185154
|
PCP4 (1 of many)
|
Purkinje cell protein 4 |
| chr20_+_30445971 | 1.50 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr2_-_22152797 | 1.32 |
ENSDART00000145188
|
cyp7a1
|
cytochrome P450, family 7, subfamily A, polypeptide 1 |
| chr15_-_12545683 | 1.24 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr13_+_27073901 | 1.20 |
ENSDART00000146227
|
slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
| chr8_-_25329967 | 1.18 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr14_-_2196267 | 1.17 |
ENSDART00000161674
ENSDART00000125674 |
pcdh2ab8
pcdh2ab9
|
protocadherin 2 alpha b 8 protocadherin 2 alpha b 9 |
| chr17_+_3128273 | 1.08 |
ENSDART00000122453
|
zgc:136872
|
zgc:136872 |
| chr17_+_3993772 | 1.05 |
ENSDART00000170822
ENSDART00000168613 ENSDART00000186174 |
tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr21_-_4032650 | 1.03 |
ENSDART00000151648
|
ntng2b
|
netrin g2b |
| chr25_-_12788370 | 1.02 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr20_+_43083745 | 0.99 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr17_-_20897407 | 0.92 |
ENSDART00000149481
|
ank3b
|
ankyrin 3b |
| chr9_-_11560110 | 0.88 |
ENSDART00000176941
|
cryba2b
|
crystallin, beta A2b |
| chr24_-_5932982 | 0.84 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr2_+_37110504 | 0.84 |
ENSDART00000132794
ENSDART00000042974 |
slc1a8b
|
solute carrier family 1 (glutamate transporter), member 8b |
| chr21_-_37889727 | 0.84 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr14_-_2036604 | 0.83 |
ENSDART00000192446
|
BX005294.2
|
|
| chr23_-_7826849 | 0.77 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr24_-_6546479 | 0.77 |
ENSDART00000160538
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr19_-_13774502 | 0.76 |
ENSDART00000159711
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr10_+_21677058 | 0.75 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr21_-_40782393 | 0.73 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr13_-_9895564 | 0.73 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr17_-_21993620 | 0.73 |
ENSDART00000179905
ENSDART00000078873 |
slc22a7b.3
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 3 |
| chr8_+_26859639 | 0.73 |
ENSDART00000133440
|
prdm2a
|
PR domain containing 2, with ZNF domain a |
| chr25_+_35189555 | 0.72 |
ENSDART00000044453
|
ano5a
|
anoctamin 5a |
| chr14_-_2358774 | 0.69 |
ENSDART00000164809
|
si:ch73-233f7.5
|
si:ch73-233f7.5 |
| chr14_-_2193248 | 0.66 |
ENSDART00000128659
|
pcdh2ab10
|
protocadherin 2 alpha b 10 |
| chr22_+_8753092 | 0.66 |
ENSDART00000140720
|
si:dkey-182g1.2
|
si:dkey-182g1.2 |
| chr17_-_20897250 | 0.66 |
ENSDART00000088106
|
ank3b
|
ankyrin 3b |
| chr17_+_13664442 | 0.65 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr7_+_8324506 | 0.64 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr19_+_42469058 | 0.64 |
ENSDART00000076915
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr17_+_47116500 | 0.63 |
ENSDART00000186627
|
CLIP4
|
si:dkeyp-47f9.4 |
| chr24_+_5840258 | 0.62 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr20_-_4031475 | 0.62 |
ENSDART00000112053
|
fam89a
|
family with sequence similarity 89, member A |
| chr4_+_10366532 | 0.61 |
ENSDART00000189901
|
kcnd2
|
potassium voltage-gated channel, Shal-related subfamily, member 2 |
| chr3_+_15773991 | 0.60 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr1_-_54706039 | 0.57 |
ENSDART00000083633
|
exosc1
|
exosome component 1 |
| chr22_+_3045495 | 0.56 |
ENSDART00000164061
|
LO017843.1
|
|
| chr2_-_48171112 | 0.55 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr21_-_22474362 | 0.55 |
ENSDART00000169659
|
myo5b
|
myosin VB |
| chr1_-_17675037 | 0.55 |
ENSDART00000142689
|
si:dkey-256e7.5
|
si:dkey-256e7.5 |
| chr6_-_51101834 | 0.54 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr23_+_39963599 | 0.53 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr21_+_36774542 | 0.51 |
ENSDART00000130790
|
prr7
|
proline rich 7 (synaptic) |
| chr20_-_40451115 | 0.51 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr7_+_14632157 | 0.50 |
ENSDART00000161264
|
ntrk3b
|
neurotrophic tyrosine kinase, receptor, type 3b |
| chr5_-_37117778 | 0.50 |
ENSDART00000149138
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr10_+_16584382 | 0.49 |
ENSDART00000112039
|
CR790388.1
|
|
| chr1_-_50438247 | 0.47 |
ENSDART00000114098
|
dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr5_+_42124706 | 0.47 |
ENSDART00000020044
ENSDART00000156372 |
shpk
|
sedoheptulokinase |
| chr24_-_11908115 | 0.47 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_-_6281503 | 0.47 |
ENSDART00000184977
|
BX004785.2
|
|
| chr10_+_45128375 | 0.47 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr17_-_6514962 | 0.46 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr22_+_26804197 | 0.45 |
ENSDART00000135688
|
si:dkey-44g23.2
|
si:dkey-44g23.2 |
| chr10_-_25817854 | 0.44 |
ENSDART00000147677
|
postna
|
periostin, osteoblast specific factor a |
| chr9_+_33009284 | 0.43 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr9_+_19039608 | 0.43 |
ENSDART00000055866
|
chmp2ba
|
charged multivesicular body protein 2Ba |
| chr22_+_5103349 | 0.42 |
ENSDART00000083474
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr5_+_43470544 | 0.41 |
ENSDART00000111587
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr8_-_50525360 | 0.40 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr9_-_48864942 | 0.40 |
ENSDART00000113855
ENSDART00000189064 |
nostrin
|
nitric oxide synthase trafficking |
| chr24_+_11105786 | 0.39 |
ENSDART00000175182
|
prlh2
|
prolactin releasing hormone 2 |
| chr3_-_32472155 | 0.39 |
ENSDART00000156280
|
si:ch211-195b15.7
|
si:ch211-195b15.7 |
| chr8_+_4838114 | 0.39 |
ENSDART00000146667
|
es1
|
es1 protein |
| chr2_-_48171441 | 0.38 |
ENSDART00000123040
|
pfkpb
|
phosphofructokinase, platelet b |
| chr9_+_21993092 | 0.37 |
ENSDART00000059693
|
crygm7
|
crystallin, gamma M7 |
| chr16_-_31756859 | 0.37 |
ENSDART00000149170
ENSDART00000126617 ENSDART00000182722 |
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_+_25651720 | 0.35 |
ENSDART00000100693
ENSDART00000100717 |
acot16
|
acyl-CoA thioesterase 16 |
| chr19_-_38830582 | 0.35 |
ENSDART00000189966
ENSDART00000183055 |
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr4_-_1908179 | 0.35 |
ENSDART00000139586
|
ano6
|
anoctamin 6 |
| chr5_-_16996482 | 0.34 |
ENSDART00000144501
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr17_-_6076084 | 0.34 |
ENSDART00000058890
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr4_-_3064101 | 0.34 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_-_35488180 | 0.32 |
ENSDART00000183258
|
rgs8
|
regulator of G protein signaling 8 |
| chr4_+_34369818 | 0.31 |
ENSDART00000160548
|
si:ch211-246b8.1
|
si:ch211-246b8.1 |
| chr5_-_30984010 | 0.30 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr10_-_25816558 | 0.30 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr8_+_14158021 | 0.29 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr24_+_36636208 | 0.29 |
ENSDART00000139211
|
si:ch73-334d15.4
|
si:ch73-334d15.4 |
| chr4_-_2036620 | 0.28 |
ENSDART00000150490
|
si:dkey-97m3.1
|
si:dkey-97m3.1 |
| chr5_-_28149767 | 0.28 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr3_+_19914332 | 0.27 |
ENSDART00000078982
|
vat1
|
vesicle amine transport 1 |
| chr12_+_30046320 | 0.27 |
ENSDART00000179904
ENSDART00000153394 |
ablim1b
|
actin binding LIM protein 1b |
| chr17_-_14591764 | 0.27 |
ENSDART00000174703
|
daam1a
|
dishevelled associated activator of morphogenesis 1a |
| chr10_+_21801030 | 0.27 |
ENSDART00000160881
ENSDART00000187969 |
pcdh1g30
|
protocadherin 1 gamma 30 |
| chr17_-_6076266 | 0.26 |
ENSDART00000171084
|
ephx2
|
epoxide hydrolase 2, cytoplasmic |
| chr6_-_57938043 | 0.26 |
ENSDART00000171073
|
tox2
|
TOX high mobility group box family member 2 |
| chr7_-_6432854 | 0.26 |
ENSDART00000172954
|
si:ch1073-153i20.4
|
si:ch1073-153i20.4 |
| chr21_+_10790680 | 0.26 |
ENSDART00000144460
|
znf532
|
zinc finger protein 532 |
| chr8_-_4097722 | 0.26 |
ENSDART00000135006
|
cux2b
|
cut-like homeobox 2b |
| chr16_+_19014886 | 0.26 |
ENSDART00000079298
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr2_-_55903520 | 0.25 |
ENSDART00000128828
|
cplx4c
|
complexin 4c |
| chr4_-_67800414 | 0.25 |
ENSDART00000160213
|
si:ch211-66c13.1
|
si:ch211-66c13.1 |
| chr9_-_32837860 | 0.24 |
ENSDART00000142227
|
mxe
|
myxovirus (influenza virus) resistance E |
| chr9_+_46745348 | 0.24 |
ENSDART00000025256
|
igfbp2b
|
insulin-like growth factor binding protein 2b |
| chr2_-_44038698 | 0.23 |
ENSDART00000079582
ENSDART00000146804 |
kirrel1b
|
kirre like nephrin family adhesion molecule 1b |
| chr14_+_22293787 | 0.23 |
ENSDART00000157967
|
atp10b
|
ATPase phospholipid transporting 10B |
| chr5_+_44064764 | 0.23 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr2_+_47623202 | 0.23 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr4_-_51460013 | 0.23 |
ENSDART00000193382
|
CR628395.1
|
|
| chr1_-_50293946 | 0.23 |
ENSDART00000053028
ENSDART00000125099 |
tbck
|
TBC1 domain containing kinase |
| chr5_-_29238889 | 0.22 |
ENSDART00000143098
|
si:dkey-61l1.4
|
si:dkey-61l1.4 |
| chr10_-_26744131 | 0.22 |
ENSDART00000020096
ENSDART00000162710 ENSDART00000179853 |
fgf13b
|
fibroblast growth factor 13b |
| chr4_-_760560 | 0.21 |
ENSDART00000103601
|
agbl5
|
ATP/GTP binding protein-like 5 |
| chr20_-_51917771 | 0.20 |
ENSDART00000147240
|
dusp10
|
dual specificity phosphatase 10 |
| chr7_-_29223614 | 0.20 |
ENSDART00000173598
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr17_+_14711765 | 0.20 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr12_+_24561832 | 0.19 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr20_+_36234335 | 0.18 |
ENSDART00000193484
ENSDART00000181664 |
cnih3
|
cornichon family AMPA receptor auxiliary protein 3 |
| chr3_+_41558682 | 0.18 |
ENSDART00000157023
|
card11
|
caspase recruitment domain family, member 11 |
| chr20_+_28266892 | 0.18 |
ENSDART00000103330
|
chac1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr1_-_26023678 | 0.18 |
ENSDART00000054202
|
si:ch211-145b13.5
|
si:ch211-145b13.5 |
| chr20_+_22666548 | 0.17 |
ENSDART00000147520
|
lnx1
|
ligand of numb-protein X 1 |
| chr15_-_17190540 | 0.17 |
ENSDART00000154801
|
itgae.2
|
integrin, alpha E, tandem duplicate 2 |
| chr8_-_27656765 | 0.16 |
ENSDART00000078491
|
mov10b.2
|
Moloney leukemia virus 10b, tandem duplicate 2 |
| chr23_+_42819221 | 0.15 |
ENSDART00000180495
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr22_+_19486711 | 0.15 |
ENSDART00000144389
ENSDART00000139256 ENSDART00000160855 |
si:dkey-78l4.10
|
si:dkey-78l4.10 |
| chr16_+_1353894 | 0.15 |
ENSDART00000148426
|
celf3b
|
cugbp, Elav-like family member 3b |
| chr2_-_59376735 | 0.15 |
ENSDART00000193624
|
ftr38
|
finTRIM family, member 38 |
| chr2_+_37975026 | 0.14 |
ENSDART00000034802
|
si:rp71-1g18.13
|
si:rp71-1g18.13 |
| chr4_+_16827390 | 0.14 |
ENSDART00000113344
|
kiss2
|
kisspeptin 2 |
| chr24_+_26017094 | 0.14 |
ENSDART00000137851
|
tfr1b
|
transferrin receptor 1b |
| chr14_-_2004291 | 0.14 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr5_-_23696926 | 0.13 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr12_+_27068525 | 0.13 |
ENSDART00000188634
|
srcap
|
Snf2-related CREBBP activator protein |
| chr18_+_18861359 | 0.12 |
ENSDART00000144605
|
pllp
|
plasmolipin |
| chr20_-_20613584 | 0.12 |
ENSDART00000140991
|
si:ch211-253p18.2
|
si:ch211-253p18.2 |
| chr7_+_29890292 | 0.12 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr10_-_11261565 | 0.11 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr7_-_34329527 | 0.11 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr3_-_59981162 | 0.10 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr20_-_11178022 | 0.10 |
ENSDART00000152246
|
flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr15_-_19677511 | 0.10 |
ENSDART00000043743
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr20_+_23315242 | 0.09 |
ENSDART00000132248
|
fryl
|
furry homolog, like |
| chr6_-_55354004 | 0.09 |
ENSDART00000165911
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr3_+_58119571 | 0.09 |
ENSDART00000108979
|
si:ch211-256e16.4
|
si:ch211-256e16.4 |
| chr1_-_38709551 | 0.09 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr7_-_4315859 | 0.09 |
ENSDART00000172762
|
si:ch211-63p21.8
|
si:ch211-63p21.8 |
| chr11_+_42587900 | 0.09 |
ENSDART00000167529
|
asb14a
|
ankyrin repeat and SOCS box containing 14a |
| chr7_-_45990386 | 0.09 |
ENSDART00000186008
|
si:ch211-260e23.7
|
si:ch211-260e23.7 |
| chr18_+_7204378 | 0.08 |
ENSDART00000142905
|
vwf
|
von Willebrand factor |
| chr12_+_48204891 | 0.08 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr16_-_12097558 | 0.08 |
ENSDART00000123142
|
pex5
|
peroxisomal biogenesis factor 5 |
| chr16_-_24612871 | 0.08 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr5_+_8919698 | 0.08 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr9_-_56272465 | 0.08 |
ENSDART00000039235
|
lcp1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr4_+_47174366 | 0.06 |
ENSDART00000170732
|
si:dkey-26m3.1
|
si:dkey-26m3.1 |
| chr2_+_36691771 | 0.06 |
ENSDART00000131978
|
ptx3b
|
pentraxin 3, long b |
| chr17_+_11500628 | 0.06 |
ENSDART00000155660
|
efcab2
|
EF-hand calcium binding domain 2 |
| chr3_-_8510201 | 0.06 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr17_-_23416897 | 0.06 |
ENSDART00000163391
|
si:ch211-149k12.3
|
si:ch211-149k12.3 |
| chr4_-_14531687 | 0.06 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr9_-_37613792 | 0.05 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr24_+_11334733 | 0.04 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr7_-_55454406 | 0.04 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr5_-_41996414 | 0.04 |
ENSDART00000143573
|
ncor1
|
nuclear receptor corepressor 1 |
| chr9_-_35875051 | 0.03 |
ENSDART00000013432
|
dnajc28
|
DnaJ (Hsp40) homolog, subfamily C, member 28 |
| chr1_-_16665044 | 0.03 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr4_-_2525916 | 0.03 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr14_+_1240604 | 0.03 |
ENSDART00000188258
|
adad1
|
adenosine deaminase domain containing 1 (testis-specific) |
| chr10_+_4046448 | 0.03 |
ENSDART00000123086
ENSDART00000052268 |
pitpnb
|
phosphatidylinositol transfer protein, beta |
| chr11_+_44226200 | 0.02 |
ENSDART00000191417
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
| chr24_-_6345647 | 0.02 |
ENSDART00000108994
|
zgc:174877
|
zgc:174877 |
| chr7_-_22632690 | 0.02 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr13_+_4440869 | 0.01 |
ENSDART00000156323
|
tbxtb
|
T-box transcription factor Tb |
| chr14_-_6943934 | 0.01 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr8_-_2153147 | 0.01 |
ENSDART00000124093
|
si:dkeyp-117b11.1
|
si:dkeyp-117b11.1 |
| chr8_+_35356944 | 0.00 |
ENSDART00000143243
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.3 | 1.0 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 1.3 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.2 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.7 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 0.8 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.5 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.1 | 0.3 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.1 | 0.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.9 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.4 | GO:0061440 | renal system vasculature development(GO:0061437) kidney vasculature development(GO:0061440) glomerulus vasculature development(GO:0072012) |
| 0.1 | 0.4 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.4 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.4 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.5 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 3.1 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.0 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.2 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 3.7 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.8 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.0 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.4 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 0.2 | GO:0039022 | pronephric duct development(GO:0039022) |
| 0.0 | 0.1 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 1.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.4 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 1.0 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.2 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 1.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.3 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.4 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 3.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 1.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.2 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 3.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID EPO PATHWAY | EPO signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.5 | 3.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.2 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |