Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
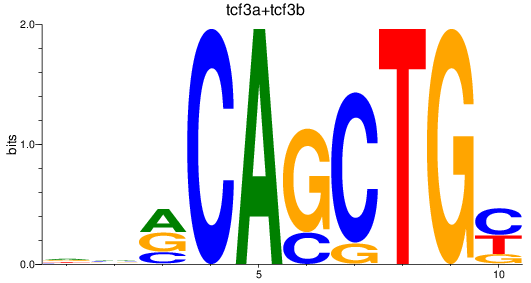
Results for tcf3a+tcf3b
Z-value: 1.06
Transcription factors associated with tcf3a+tcf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf3a
|
ENSDARG00000005915 | transcription factor 3a |
|
tcf3b
|
ENSDARG00000099999 | transcription factor 3b |
|
tcf3b
|
ENSDARG00000112646 | transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf3b | dr11_v1_chr22_-_20342260_20342260 | 0.74 | 2.9e-04 | Click! |
| tcf3a | dr11_v1_chr2_-_57378748_57378748 | 0.30 | 2.1e-01 | Click! |
Activity profile of tcf3a+tcf3b motif
Sorted Z-values of tcf3a+tcf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_29962345 | 3.40 |
ENSDART00000136819
|
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr5_-_14521500 | 2.92 |
ENSDART00000176565
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr19_+_15440841 | 2.58 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_15441022 | 2.57 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr22_-_10541372 | 2.38 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr5_+_46424437 | 2.22 |
ENSDART00000186511
|
vcana
|
versican a |
| chr14_-_30390145 | 1.99 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr7_+_2455344 | 1.89 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr24_-_9300160 | 1.89 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr21_-_19919020 | 1.84 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr17_-_14836320 | 1.80 |
ENSDART00000157051
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr22_-_10541712 | 1.75 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr21_-_23331619 | 1.71 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr3_+_14641962 | 1.69 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr15_+_7187228 | 1.68 |
ENSDART00000109394
|
her13
|
hairy-related 13 |
| chr10_+_31244619 | 1.64 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr14_-_26482096 | 1.62 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr13_-_31938512 | 1.60 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr17_+_27434626 | 1.59 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr13_+_11439486 | 1.59 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr2_-_17114852 | 1.58 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr7_-_18881358 | 1.56 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr6_-_11073880 | 1.54 |
ENSDART00000035905
|
sumo3a
|
small ubiquitin-like modifier 3a |
| chr21_-_21465111 | 1.52 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr17_-_14815557 | 1.52 |
ENSDART00000154473
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr17_+_132555 | 1.49 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr18_-_16922905 | 1.48 |
ENSDART00000187165
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr1_-_156375 | 1.46 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr20_-_20930926 | 1.40 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr15_-_5624361 | 1.39 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr6_+_40629066 | 1.39 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr20_+_43691208 | 1.36 |
ENSDART00000152976
ENSDART00000045185 |
lin9
|
lin-9 DREAM MuvB core complex component |
| chr11_-_17755444 | 1.36 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr8_-_18582922 | 1.36 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr21_-_45882643 | 1.35 |
ENSDART00000168703
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr16_+_46294337 | 1.35 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr9_+_28232522 | 1.35 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr8_+_12118097 | 1.34 |
ENSDART00000081819
|
endog
|
endonuclease G |
| chr22_+_3914318 | 1.32 |
ENSDART00000188774
ENSDART00000082034 |
FO904903.1
|
Danio rerio major histocompatibility complex class I ULA (mhc1ula), mRNA. |
| chr15_-_4528326 | 1.32 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr13_+_18321140 | 1.32 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr14_+_32964 | 1.31 |
ENSDART00000166173
|
lyar
|
Ly1 antibody reactive homolog (mouse) |
| chr14_-_25928899 | 1.29 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr19_-_10214264 | 1.28 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr14_-_25930182 | 1.27 |
ENSDART00000018651
ENSDART00000147991 |
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr24_+_39108243 | 1.26 |
ENSDART00000156353
|
mss51
|
MSS51 mitochondrial translational activator |
| chr21_-_44772738 | 1.26 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr8_+_22405477 | 1.26 |
ENSDART00000148267
|
si:dkey-23c22.7
|
si:dkey-23c22.7 |
| chr3_+_26245731 | 1.25 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr8_-_47844456 | 1.23 |
ENSDART00000145429
|
si:dkeyp-104h9.5
|
si:dkeyp-104h9.5 |
| chr5_+_32835219 | 1.22 |
ENSDART00000140832
ENSDART00000186055 |
si:ch211-208h16.4
|
si:ch211-208h16.4 |
| chr8_-_44298964 | 1.21 |
ENSDART00000098520
|
fzd10
|
frizzled class receptor 10 |
| chr8_+_46327350 | 1.21 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr12_+_26632448 | 1.21 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr16_-_29164379 | 1.19 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr23_+_26079467 | 1.18 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr8_+_39570615 | 1.17 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr4_-_76488581 | 1.17 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr3_+_59935606 | 1.16 |
ENSDART00000154157
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr15_-_23529945 | 1.15 |
ENSDART00000152543
|
hmbsb
|
hydroxymethylbilane synthase, b |
| chr23_+_21459263 | 1.15 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr11_-_18705303 | 1.14 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr22_+_465269 | 1.14 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr21_+_39941559 | 1.14 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr12_+_9499993 | 1.14 |
ENSDART00000135871
|
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr23_-_27607039 | 1.13 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr13_+_51579851 | 1.13 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr8_-_13735572 | 1.12 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr19_+_48359259 | 1.12 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr13_-_21672131 | 1.12 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr3_+_60716904 | 1.12 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr7_+_49715750 | 1.11 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr1_+_227241 | 1.11 |
ENSDART00000003317
|
tfdp1b
|
transcription factor Dp-1, b |
| chr20_+_1316495 | 1.10 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr15_-_41333064 | 1.08 |
ENSDART00000144678
|
si:dkey-121n8.7
|
si:dkey-121n8.7 |
| chr8_-_50979047 | 1.08 |
ENSDART00000184788
ENSDART00000180906 |
zgc:91909
|
zgc:91909 |
| chr9_+_3429662 | 1.08 |
ENSDART00000160977
ENSDART00000114168 ENSDART00000082153 |
CU469503.1
itga6a
|
integrin, alpha 6a |
| chr11_-_308838 | 1.06 |
ENSDART00000112538
|
poc1a
|
POC1 centriolar protein A |
| chr2_+_36701322 | 1.06 |
ENSDART00000002510
|
golim4b
|
golgi integral membrane protein 4b |
| chr21_-_22357985 | 1.06 |
ENSDART00000101751
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr7_+_19552381 | 1.05 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr6_-_18618106 | 1.05 |
ENSDART00000161562
|
znf207b
|
zinc finger protein 207, b |
| chr2_+_20793982 | 1.05 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr5_-_30475011 | 1.04 |
ENSDART00000187501
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr13_+_1131748 | 1.04 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr13_+_28612313 | 1.04 |
ENSDART00000077383
|
borcs7
|
BLOC-1 related complex subunit 7 |
| chr12_+_1592146 | 1.03 |
ENSDART00000184575
ENSDART00000192902 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr5_-_8164439 | 1.03 |
ENSDART00000189912
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr20_+_4221978 | 1.03 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr16_+_28728347 | 1.03 |
ENSDART00000149240
|
si:dkey-24i24.3
|
si:dkey-24i24.3 |
| chr17_+_8799451 | 1.02 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_-_1051438 | 1.02 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr5_-_63515210 | 1.02 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr25_+_3294150 | 1.02 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr9_-_7390388 | 1.00 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr9_-_27717006 | 1.00 |
ENSDART00000146860
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr9_+_32872690 | 1.00 |
ENSDART00000020798
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr2_-_17115256 | 0.99 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr17_-_9930163 | 0.99 |
ENSDART00000149640
|
egln3
|
egl-9 family hypoxia-inducible factor 3 |
| chr10_-_1961930 | 0.99 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr5_-_22052852 | 0.99 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr5_+_13549603 | 0.99 |
ENSDART00000171270
|
ckap2l
|
cytoskeleton associated protein 2-like |
| chr18_+_30441740 | 0.99 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr16_+_19537073 | 0.98 |
ENSDART00000190590
|
sp8b
|
sp8 transcription factor b |
| chr7_-_18470963 | 0.98 |
ENSDART00000173929
ENSDART00000173638 |
znf16l
|
zinc finger protein 16 like |
| chr16_+_54875530 | 0.98 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr14_-_9281232 | 0.98 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr23_+_28077953 | 0.98 |
ENSDART00000186122
ENSDART00000111570 |
slc26a10
|
solute carrier family 26, member 10 |
| chr16_+_26706519 | 0.97 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr11_+_3501669 | 0.97 |
ENSDART00000160808
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr8_-_32805214 | 0.97 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr23_-_19831739 | 0.96 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr11_-_41966854 | 0.96 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr12_+_9499742 | 0.95 |
ENSDART00000044150
ENSDART00000136354 |
dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr22_+_22438783 | 0.95 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr13_+_40770628 | 0.95 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr1_+_8694196 | 0.95 |
ENSDART00000025604
|
zgc:77849
|
zgc:77849 |
| chr17_+_8799661 | 0.94 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr19_-_48010490 | 0.94 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr14_+_21828993 | 0.94 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr3_+_40164129 | 0.94 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr3_-_40275096 | 0.94 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr6_+_49052741 | 0.93 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr1_-_53880639 | 0.93 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr11_-_44898129 | 0.93 |
ENSDART00000157615
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr19_-_21766461 | 0.92 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr21_-_26677834 | 0.92 |
ENSDART00000077381
|
nxf1
|
nuclear RNA export factor 1 |
| chr1_+_38142354 | 0.92 |
ENSDART00000179352
|
galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_+_37838259 | 0.92 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr8_+_15254564 | 0.92 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr9_+_41024973 | 0.92 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr7_-_50367642 | 0.92 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr21_-_30026359 | 0.92 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr2_+_42294944 | 0.91 |
ENSDART00000140599
ENSDART00000178811 |
ftr06
|
finTRIM family, member 6 |
| chr15_+_29472065 | 0.91 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr24_+_25210015 | 0.91 |
ENSDART00000081043
|
cip2a
|
cell proliferation regulating inhibitor of protein phosphatase 2A |
| chr6_-_36552844 | 0.91 |
ENSDART00000023613
|
her6
|
hairy-related 6 |
| chr13_+_36595618 | 0.91 |
ENSDART00000022684
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr9_-_52598343 | 0.91 |
ENSDART00000167922
|
xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr3_-_26978793 | 0.91 |
ENSDART00000155396
|
nubp1
|
nucleotide binding protein 1 (MinD homolog, E. coli) |
| chr16_-_34212912 | 0.91 |
ENSDART00000145017
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr8_-_50287949 | 0.90 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr13_+_2894536 | 0.90 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr13_+_25200105 | 0.89 |
ENSDART00000039640
|
ap3m1
|
adaptor-related protein complex 3, mu 1 subunit |
| chr7_+_18878973 | 0.89 |
ENSDART00000037846
|
focad
|
focadhesin |
| chr8_-_32346300 | 0.89 |
ENSDART00000145325
|
ipo11
|
importin 11 |
| chr2_+_27386617 | 0.89 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr12_-_48168135 | 0.89 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr24_-_9294134 | 0.89 |
ENSDART00000082434
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr18_-_20444296 | 0.89 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr2_-_24402341 | 0.89 |
ENSDART00000155442
ENSDART00000088572 |
zgc:154006
|
zgc:154006 |
| chr6_+_59991076 | 0.88 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr16_+_19536614 | 0.87 |
ENSDART00000112894
ENSDART00000079201 ENSDART00000139357 |
sp8b
|
sp8 transcription factor b |
| chr21_+_5192016 | 0.87 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr2_-_42628028 | 0.87 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr4_-_2052687 | 0.87 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr17_+_24809743 | 0.86 |
ENSDART00000135986
|
spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr25_+_5249513 | 0.86 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr20_+_35247854 | 0.86 |
ENSDART00000184124
|
fbxo16
|
F-box protein 16 |
| chr19_-_2317558 | 0.86 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr20_+_46274118 | 0.86 |
ENSDART00000124961
|
taar14g
|
trace amine associated receptor 14g |
| chr23_-_10696626 | 0.86 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr4_-_76488854 | 0.86 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr19_-_27564458 | 0.85 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr19_-_30810328 | 0.85 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr14_+_52481288 | 0.85 |
ENSDART00000169164
ENSDART00000159297 |
tcerg1a
|
transcription elongation regulator 1a (CA150) |
| chr6_-_10728057 | 0.84 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr13_+_22295905 | 0.84 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr11_-_18015534 | 0.84 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr12_+_3262564 | 0.84 |
ENSDART00000184264
|
tmem101
|
transmembrane protein 101 |
| chr20_-_9194257 | 0.84 |
ENSDART00000133012
|
ylpm1
|
YLP motif containing 1 |
| chr18_-_6862738 | 0.84 |
ENSDART00000192592
|
CT009745.1
|
|
| chr13_-_46200240 | 0.84 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr8_-_1255321 | 0.84 |
ENSDART00000149605
|
cdc14b
|
cell division cycle 14B |
| chr19_-_4793263 | 0.84 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr22_+_22021936 | 0.83 |
ENSDART00000149586
|
gna15.1
|
guanine nucleotide binding protein (G protein), alpha 15 (Gq class), tandem duplicate 1 |
| chr21_+_39941875 | 0.83 |
ENSDART00000190414
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr19_-_17972734 | 0.83 |
ENSDART00000126298
|
ints8
|
integrator complex subunit 8 |
| chr8_-_11229523 | 0.83 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr24_+_4978055 | 0.82 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr6_+_40775800 | 0.82 |
ENSDART00000085090
|
si:ch211-157b11.8
|
si:ch211-157b11.8 |
| chr5_+_34549845 | 0.81 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr16_-_29146624 | 0.81 |
ENSDART00000159814
ENSDART00000009826 |
mef2d
|
myocyte enhancer factor 2d |
| chr17_+_38262408 | 0.81 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr23_-_32236122 | 0.81 |
ENSDART00000103343
|
grasp
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr18_+_22220656 | 0.81 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr17_+_16090436 | 0.80 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr17_-_49407091 | 0.80 |
ENSDART00000021950
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr5_+_22098591 | 0.80 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr24_+_4977862 | 0.80 |
ENSDART00000114537
|
zic4
|
zic family member 4 |
| chr8_-_37043900 | 0.79 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr23_+_38245610 | 0.79 |
ENSDART00000191386
|
znf217
|
zinc finger protein 217 |
| chr17_-_27419319 | 0.79 |
ENSDART00000127043
|
ythdf2
|
YTH N(6)-methyladenosine RNA binding protein 2 |
| chr1_+_260039 | 0.79 |
ENSDART00000092584
ENSDART00000111806 |
cenpe
|
centromere protein E |
| chr23_+_44236281 | 0.79 |
ENSDART00000149842
|
MEPCE
|
si:ch1073-157b13.1 |
| chr19_+_42432625 | 0.79 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr19_+_29808699 | 0.78 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr7_+_756942 | 0.78 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr19_+_15485556 | 0.78 |
ENSDART00000079014
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr20_-_37831849 | 0.77 |
ENSDART00000188483
ENSDART00000153005 ENSDART00000142364 |
si:ch211-147d7.5
|
si:ch211-147d7.5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf3a+tcf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.7 | 2.2 | GO:0001977 | renal system process involved in regulation of blood volume(GO:0001977) renal system process involved in regulation of systemic arterial blood pressure(GO:0003071) regulation of glomerular filtration(GO:0003093) |
| 0.6 | 1.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.6 | 1.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.6 | 1.7 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.5 | 1.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.5 | 1.5 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.4 | 1.2 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.4 | 1.9 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.3 | 2.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 1.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.3 | 1.0 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.3 | 2.0 | GO:0043091 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.3 | 1.0 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.3 | 0.9 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.3 | 0.9 | GO:0051026 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.3 | 0.9 | GO:1990120 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.3 | 1.1 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.3 | 1.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 1.2 | GO:0043393 | regulation of protein binding(GO:0043393) |
| 0.2 | 0.7 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 1.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.2 | 2.9 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 | 3.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 1.2 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.2 | 3.0 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.2 | 1.6 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 0.7 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.2 | 0.9 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 0.7 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 0.7 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.4 | GO:0030910 | olfactory placode formation(GO:0030910) |
| 0.2 | 2.4 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.2 | 2.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 1.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 1.4 | GO:0070601 | meiotic sister chromatid cohesion, centromeric(GO:0051754) centromeric sister chromatid cohesion(GO:0070601) |
| 0.2 | 0.6 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 0.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.2 | 1.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 0.7 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.4 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.2 | 0.9 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 1.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.2 | 0.5 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.5 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.7 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.2 | 1.3 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.2 | 0.5 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.2 | 1.0 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 1.0 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 0.6 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.2 | 0.6 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 1.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 0.5 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.2 | 0.8 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.2 | 0.6 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.6 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.7 | GO:0016037 | light absorption(GO:0016037) |
| 0.1 | 0.4 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.1 | 0.7 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.4 | GO:0032369 | negative regulation of lipid transport(GO:0032369) negative regulation of lipid catabolic process(GO:0050995) |
| 0.1 | 0.5 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.9 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 0.9 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.8 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.4 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.7 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.4 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 1.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.6 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.1 | 0.4 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.2 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 1.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.6 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 2.7 | GO:2000826 | regulation of heart morphogenesis(GO:2000826) |
| 0.1 | 0.6 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.7 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 0.4 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.6 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 1.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.3 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.1 | 0.7 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.5 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.4 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.1 | 2.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.5 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.9 | GO:0021794 | thalamus development(GO:0021794) |
| 0.1 | 0.5 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.7 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 1.2 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 1.9 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 3.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 0.7 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0021855 | axon target recognition(GO:0007412) hypothalamus cell migration(GO:0021855) |
| 0.1 | 0.8 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 | 2.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.5 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 1.4 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 0.4 | GO:0014856 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 1.1 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.1 | 0.5 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 | 0.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.1 | 1.6 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.6 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 0.6 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.4 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 0.9 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.2 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.7 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.1 | 0.4 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.1 | 0.4 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 0.2 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.6 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.2 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.1 | 0.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.2 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.1 | 0.8 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.1 | 1.5 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.5 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.7 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.1 | GO:0051148 | negative regulation of muscle cell differentiation(GO:0051148) |
| 0.1 | 0.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.4 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 0.6 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.3 | GO:0098921 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 0.2 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.4 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 0.5 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.4 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.8 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 1.7 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.0 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.1 | 1.8 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.3 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 0.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.4 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.3 | GO:0060585 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.6 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.4 | GO:1902229 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902229) |
| 0.1 | 0.1 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:1990564 | IRE1-mediated unfolded protein response(GO:0036498) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.2 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.2 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 1.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 5.6 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 0.4 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 0.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.2 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.1 | 0.6 | GO:0060035 | notochord cell development(GO:0060035) |
| 0.1 | 0.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.4 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.2 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.5 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.5 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 1.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.3 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.2 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 0.7 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 1.3 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 1.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.9 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.5 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.4 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:2000320 | regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 1.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 1.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.7 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.0 | 0.4 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.4 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 1.7 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 1.8 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.0 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.3 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.0 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.8 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.4 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.9 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.6 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 1.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 1.0 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.6 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 1.1 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.1 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.6 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.5 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.5 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.2 | GO:0035909 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.0 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0043388 | positive regulation of DNA binding(GO:0043388) positive regulation of binding(GO:0051099) |
| 0.0 | 0.1 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.7 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.7 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.4 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.4 | GO:0033198 | response to ATP(GO:0033198) |
| 0.0 | 0.6 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 1.1 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:1903352 | mitochondrial ornithine transport(GO:0000066) ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 1.3 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.1 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.6 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.2 | GO:0086002 | cardiac muscle cell action potential involved in contraction(GO:0086002) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 1.1 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 2.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.2 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.2 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.7 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 1.1 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.3 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.6 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.1 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.0 | 0.2 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.4 | GO:0035723 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.4 | GO:0033077 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.0 | 0.2 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.5 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.3 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.7 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.8 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.7 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.8 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:1903392 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.2 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 1.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.9 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.2 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) regulation of phospholipid metabolic process(GO:1903725) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.3 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.4 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.7 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.0 | GO:0021695 | cerebellar Purkinje cell layer development(GO:0021680) cerebellar cortex development(GO:0021695) |
| 0.0 | 0.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.5 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.0 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.5 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.8 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.1 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.5 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.5 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.3 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.3 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate pattern specification(GO:0060896) neural plate regionalization(GO:0060897) |
| 0.0 | 0.2 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 1.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.7 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.9 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.3 | GO:0016072 | rRNA metabolic process(GO:0016072) |
| 0.0 | 0.5 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.5 | GO:0002573 | myeloid leukocyte differentiation(GO:0002573) |
| 0.0 | 0.0 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.7 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.3 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.7 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.2 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.3 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 2.0 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.9 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.2 | 0.9 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 1.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.2 | 1.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.2 | 1.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.7 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.2 | 0.5 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.2 | 0.5 | GO:0072380 | TRC complex(GO:0072380) |
| 0.2 | 0.9 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.1 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.4 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 1.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.7 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.9 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.5 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.7 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.8 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.4 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 2.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 1.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.6 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.5 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 1.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.2 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 2.6 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 1.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.6 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.7 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.6 | GO:0070449 | elongin complex(GO:0070449) |
| 0.1 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 1.1 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.1 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 2.0 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 0.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.2 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 2.0 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.7 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 2.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.2 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 3.4 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 4.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0097519 | DNA recombinase complex(GO:0097519) |
| 0.0 | 0.8 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.5 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 1.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 2.0 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.6 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.5 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.1 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 3.0 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 5.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.6 | GO:0045495 | pole plasm(GO:0045495) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 4.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 3.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0044327 | dendritic spine head(GO:0044327) postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.4 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 75.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.0 | GO:0097541 | axonemal basal plate(GO:0097541) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.5 | 2.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 2.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 1.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.3 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.3 | 1.0 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.3 | 0.9 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.3 | 1.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 1.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 0.8 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 0.7 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.2 | 1.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.2 | 0.7 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.2 | 1.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.6 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 0.6 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 0.8 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.2 | 1.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.2 | 0.7 | GO:0019972 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.2 | 0.5 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.6 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.2 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 0.5 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.1 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 2.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.5 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.1 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 2.0 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.5 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.1 | 0.5 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.8 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.4 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 1.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.6 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 1.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.6 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 1.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.1 | 0.8 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.6 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.1 | 1.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 1.0 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.3 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.1 | 1.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.8 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 1.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 0.4 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.5 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.8 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 0.3 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.9 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.2 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 4.6 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 1.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.1 | 0.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.6 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 0.2 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 1.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.5 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.9 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.2 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 1.5 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 3.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.2 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.7 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.8 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 2.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.8 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 2.0 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 1.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 34.8 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.6 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.8 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.4 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.0 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 4.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.3 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 14.0 | GO:0003677 | DNA binding(GO:0003677) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 1.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.1 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 7.4 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.5 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.1 | 1.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 1.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.6 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.6 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 2.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.4 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.3 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 4.0 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.2 | 4.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 1.4 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 2.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.5 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.0 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 0.7 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.4 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 1.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.1 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.7 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 2.8 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.1 | 1.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.1 | 0.2 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.1 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.7 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 2.2 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.6 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.8 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.3 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME MITOTIC G2 G2 M PHASES | Genes involved in Mitotic G2-G2/M phases |
| 0.0 | 1.1 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.8 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.4 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.8 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 1.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME DEADENYLATION DEPENDENT MRNA DECAY | Genes involved in Deadenylation-dependent mRNA decay |
| 0.0 | 0.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.9 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.1 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |