Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
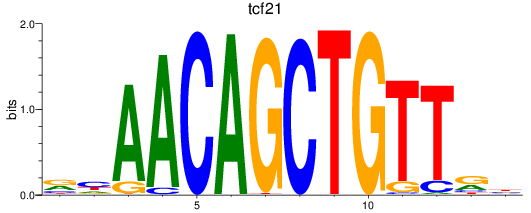
Results for tcf21
Z-value: 0.76
Transcription factors associated with tcf21
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf21
|
ENSDARG00000036869 | transcription factor 21 |
|
tcf21
|
ENSDARG00000111209 | transcription factor 21 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf21 | dr11_v1_chr23_-_31555696_31555696 | 0.28 | 2.5e-01 | Click! |
Activity profile of tcf21 motif
Sorted Z-values of tcf21 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_13256415 | 4.31 |
ENSDART00000144542
|
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr9_-_22122590 | 4.25 |
ENSDART00000108748
ENSDART00000174614 |
crygm2d9
|
crystallin, gamma M2d9 |
| chr8_-_25247284 | 4.03 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr19_+_9174166 | 3.32 |
ENSDART00000104637
ENSDART00000150968 |
si:ch211-81a5.8
|
si:ch211-81a5.8 |
| chr21_+_39185761 | 3.24 |
ENSDART00000075992
ENSDART00000140644 |
cryba1b
|
crystallin, beta A1b |
| chr21_+_39185461 | 2.92 |
ENSDART00000178419
|
cryba1b
|
crystallin, beta A1b |
| chr15_+_20403084 | 2.77 |
ENSDART00000141388
ENSDART00000152734 |
cox7a1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle) |
| chr10_+_21576909 | 2.44 |
ENSDART00000168604
ENSDART00000166533 |
pcdh1a3
|
protocadherin 1 alpha 3 |
| chr9_+_13714379 | 2.38 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr6_+_13742899 | 2.37 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr11_+_29975830 | 2.25 |
ENSDART00000148929
|
si:ch73-226l13.2
|
si:ch73-226l13.2 |
| chr1_-_22691182 | 2.12 |
ENSDART00000076766
|
fgfbp2b
|
fibroblast growth factor binding protein 2b |
| chr25_-_19655820 | 2.03 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr21_-_43952958 | 2.03 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_-_11266882 | 1.99 |
ENSDART00000020256
|
lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr1_-_14258409 | 1.98 |
ENSDART00000079359
|
pde5aa
|
phosphodiesterase 5A, cGMP-specific, a |
| chr6_-_39344259 | 1.86 |
ENSDART00000104074
|
zgc:158846
|
zgc:158846 |
| chr25_-_20258508 | 1.75 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr10_-_24362775 | 1.75 |
ENSDART00000182104
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr8_+_39724138 | 1.72 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr23_+_29908348 | 1.67 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr5_-_24238733 | 1.65 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr8_-_44004135 | 1.54 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr24_-_26383978 | 1.51 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr22_+_1049367 | 1.50 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr4_+_7508316 | 1.49 |
ENSDART00000170924
ENSDART00000170933 ENSDART00000164985 ENSDART00000167571 ENSDART00000158843 ENSDART00000158999 |
tnnt2e
|
troponin T2e, cardiac |
| chr11_+_21910343 | 1.48 |
ENSDART00000161485
|
foxp4
|
forkhead box P4 |
| chr3_+_56876280 | 1.47 |
ENSDART00000154197
|
amn
|
amnion associated transmembrane protein |
| chr9_-_49531762 | 1.46 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr2_+_6734108 | 1.45 |
ENSDART00000112227
|
brinp3b
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3b |
| chr11_-_183328 | 1.43 |
ENSDART00000168562
|
avil
|
advillin |
| chr8_-_43997538 | 1.40 |
ENSDART00000186449
|
rimbp2
|
RIMS binding protein 2 |
| chr5_+_37068223 | 1.39 |
ENSDART00000164279
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr14_+_32837914 | 1.33 |
ENSDART00000158888
|
arr3b
|
arrestin 3b, retinal (X-arrestin) |
| chr7_+_34231782 | 1.32 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr5_+_28830643 | 1.32 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr24_+_37361111 | 1.31 |
ENSDART00000078771
|
si:ch211-183d21.3
|
si:ch211-183d21.3 |
| chr18_+_39028417 | 1.29 |
ENSDART00000148428
|
myo5aa
|
myosin VAa |
| chr24_-_29906904 | 1.27 |
ENSDART00000178791
|
LO018627.1
|
|
| chr15_-_16098531 | 1.26 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr23_+_44307996 | 1.26 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr6_+_112579 | 1.26 |
ENSDART00000034505
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr23_+_20563779 | 1.24 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr7_+_26224211 | 1.23 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr13_-_40499296 | 1.22 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr18_+_907266 | 1.18 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr24_+_22056386 | 1.16 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr24_-_35707552 | 1.13 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr15_-_10455438 | 1.11 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr20_-_26551210 | 1.10 |
ENSDART00000077715
|
si:dkey-25e12.3
|
si:dkey-25e12.3 |
| chr19_+_43669122 | 1.05 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr5_+_28849155 | 1.04 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr5_-_30079434 | 1.04 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr10_+_10738880 | 1.03 |
ENSDART00000004181
|
slc27a4
|
solute carrier family 27 (fatty acid transporter), member 4 |
| chr25_+_3326885 | 1.03 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr7_+_10610791 | 1.01 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr8_+_25247245 | 1.00 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr19_-_2085027 | 0.98 |
ENSDART00000063615
|
snx13
|
sorting nexin 13 |
| chr15_-_47115787 | 0.98 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr3_+_29714775 | 0.97 |
ENSDART00000041388
|
cacng2a
|
calcium channel, voltage-dependent, gamma subunit 2a |
| chr11_+_24819624 | 0.97 |
ENSDART00000155514
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr23_-_16692312 | 0.96 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr8_+_14792830 | 0.96 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr4_-_75175407 | 0.96 |
ENSDART00000180125
|
CABZ01043953.1
|
|
| chr20_+_42668875 | 0.95 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr16_-_22004402 | 0.95 |
ENSDART00000145100
ENSDART00000188217 ENSDART00000180083 |
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr7_-_44672095 | 0.95 |
ENSDART00000073745
|
cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr3_+_22935183 | 0.95 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr20_+_46560258 | 0.94 |
ENSDART00000122115
|
si:ch211-153j24.3
|
si:ch211-153j24.3 |
| chr7_-_24047316 | 0.94 |
ENSDART00000184799
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr21_-_16632808 | 0.94 |
ENSDART00000172645
|
unc5da
|
unc-5 netrin receptor Da |
| chr21_-_293146 | 0.93 |
ENSDART00000157781
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr6_+_37752781 | 0.92 |
ENSDART00000154364
|
herc2
|
HECT and RLD domain containing E3 ubiquitin protein ligase 2 |
| chr24_-_31123365 | 0.92 |
ENSDART00000182947
|
tmem56a
|
transmembrane protein 56a |
| chr4_-_815871 | 0.91 |
ENSDART00000067455
|
dpysl5b
|
dihydropyrimidinase-like 5b |
| chr20_+_18163355 | 0.89 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr4_+_53246788 | 0.88 |
ENSDART00000184708
ENSDART00000169256 |
si:dkey-250k10.1
|
si:dkey-250k10.1 |
| chr14_+_21783229 | 0.88 |
ENSDART00000170784
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr21_+_27382893 | 0.87 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr21_-_41617372 | 0.85 |
ENSDART00000187171
|
BX005397.1
|
|
| chr18_-_399554 | 0.84 |
ENSDART00000164374
ENSDART00000186311 ENSDART00000181816 ENSDART00000181892 |
si:ch211-79l17.1
|
si:ch211-79l17.1 |
| chr6_+_58915889 | 0.84 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr17_-_14726824 | 0.83 |
ENSDART00000162947
|
si:ch73-305o9.3
|
si:ch73-305o9.3 |
| chr13_+_23677949 | 0.83 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr19_-_32804535 | 0.83 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr9_-_35155089 | 0.81 |
ENSDART00000077901
|
appb
|
amyloid beta (A4) precursor protein b |
| chr10_+_24445698 | 0.81 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr25_-_32869794 | 0.78 |
ENSDART00000162784
|
tmem266
|
transmembrane protein 266 |
| chr18_-_7948188 | 0.78 |
ENSDART00000091805
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr16_+_27957808 | 0.77 |
ENSDART00000133696
|
znf804b
|
zinc finger protein 804B |
| chr21_+_19418563 | 0.76 |
ENSDART00000181113
ENSDART00000080110 |
amacr
|
alpha-methylacyl-CoA racemase |
| chr6_+_28054639 | 0.76 |
ENSDART00000187478
ENSDART00000189194 |
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr6_-_35032792 | 0.76 |
ENSDART00000168256
|
ddr2a
|
discoidin domain receptor tyrosine kinase 2a |
| chr8_-_6943155 | 0.76 |
ENSDART00000139545
ENSDART00000033294 |
wdr13
|
WD repeat domain 13 |
| chr22_-_10470663 | 0.75 |
ENSDART00000143352
|
omd
|
osteomodulin |
| chr13_+_2442841 | 0.75 |
ENSDART00000114456
ENSDART00000137124 ENSDART00000193737 ENSDART00000189722 ENSDART00000187485 |
arfgef3
|
ARFGEF family member 3 |
| chr13_+_1575276 | 0.74 |
ENSDART00000165987
|
DST
|
dystonin |
| chr14_-_46832825 | 0.74 |
ENSDART00000149571
|
ldb2a
|
LIM domain binding 2a |
| chr16_+_47428721 | 0.73 |
ENSDART00000180597
ENSDART00000032309 |
slc25a32b
|
solute carrier family 25 (mitochondrial folate carrier), member 32b |
| chr5_+_28848870 | 0.73 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr23_+_23183449 | 0.73 |
ENSDART00000132296
|
klhl17
|
kelch-like family member 17 |
| chr21_-_22635245 | 0.72 |
ENSDART00000115224
ENSDART00000101782 |
nectin1a
|
nectin cell adhesion molecule 1a |
| chr7_-_24046999 | 0.71 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr13_-_280827 | 0.71 |
ENSDART00000144819
|
slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr17_-_19828505 | 0.71 |
ENSDART00000154745
|
fmn2a
|
formin 2a |
| chr2_+_243778 | 0.70 |
ENSDART00000182262
|
CABZ01085887.1
|
|
| chr12_+_2660733 | 0.70 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr5_-_57480660 | 0.69 |
ENSDART00000147875
ENSDART00000142776 |
si:ch211-202f5.2
|
si:ch211-202f5.2 |
| chr7_+_17096281 | 0.68 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr8_-_44868020 | 0.68 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr21_+_8427059 | 0.68 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr3_-_28075756 | 0.68 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr19_-_24758109 | 0.68 |
ENSDART00000161016
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr22_+_24157807 | 0.67 |
ENSDART00000159165
|
b3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr3_-_34724879 | 0.66 |
ENSDART00000177021
|
thraa
|
thyroid hormone receptor alpha a |
| chr1_+_25801648 | 0.65 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr5_-_38428217 | 0.63 |
ENSDART00000191902
|
pld2
|
phospholipase D2 |
| chr8_-_1838315 | 0.63 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr10_-_34741738 | 0.62 |
ENSDART00000163072
|
dclk1a
|
doublecortin-like kinase 1a |
| chr2_-_34483597 | 0.62 |
ENSDART00000133224
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr14_+_21783400 | 0.62 |
ENSDART00000164023
|
ankrd13d
|
ankyrin repeat domain 13 family, member D |
| chr8_+_40080728 | 0.62 |
ENSDART00000111506
|
lrrc75ba
|
leucine rich repeat containing 75Ba |
| chr21_-_36571804 | 0.62 |
ENSDART00000138129
|
wwc1
|
WW and C2 domain containing 1 |
| chr13_+_2625150 | 0.60 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr2_+_37480669 | 0.60 |
ENSDART00000029801
|
sppl2
|
signal peptide peptidase-like 2 |
| chr15_+_29276508 | 0.59 |
ENSDART00000170537
ENSDART00000126559 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr3_+_4266289 | 0.59 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr15_+_28096152 | 0.58 |
ENSDART00000100293
ENSDART00000140092 |
crybb1l3
|
crystallin, beta B1, like 3 |
| chr3_+_14571813 | 0.58 |
ENSDART00000146728
ENSDART00000171731 |
znf653
|
zinc finger protein 653 |
| chr18_-_10995410 | 0.58 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr25_-_36044583 | 0.58 |
ENSDART00000073432
|
rbl2
|
retinoblastoma-like 2 (p130) |
| chr7_+_6969909 | 0.57 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr8_+_36500061 | 0.57 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr17_-_51195651 | 0.56 |
ENSDART00000191205
ENSDART00000088185 |
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr8_+_53563949 | 0.55 |
ENSDART00000190022
ENSDART00000190927 |
CU861886.1
|
|
| chr14_+_12110020 | 0.55 |
ENSDART00000192462
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr18_+_23218980 | 0.55 |
ENSDART00000185014
|
mef2aa
|
myocyte enhancer factor 2aa |
| chr6_-_40098641 | 0.54 |
ENSDART00000017402
|
ip6k2b
|
inositol hexakisphosphate kinase 2b |
| chr14_+_12109535 | 0.54 |
ENSDART00000054620
ENSDART00000121913 |
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr10_-_7892192 | 0.53 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr5_+_67448664 | 0.53 |
ENSDART00000105731
|
si:dkey-251i10.1
|
si:dkey-251i10.1 |
| chr8_+_30456161 | 0.53 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr13_+_24584401 | 0.53 |
ENSDART00000057599
|
fuom
|
fucose mutarotase |
| chr14_+_9287683 | 0.52 |
ENSDART00000122485
|
msnb
|
moesin b |
| chr21_-_12749501 | 0.51 |
ENSDART00000179724
|
LO018011.1
|
|
| chr15_+_42599501 | 0.51 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr8_-_14151051 | 0.51 |
ENSDART00000090427
|
si:dkey-6n6.1
|
si:dkey-6n6.1 |
| chr13_+_21601149 | 0.51 |
ENSDART00000179369
|
sh2d4ba
|
SH2 domain containing 4Ba |
| chr23_+_25879320 | 0.50 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_+_77526 | 0.50 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr24_+_38155830 | 0.50 |
ENSDART00000152019
|
si:ch211-234p6.5
|
si:ch211-234p6.5 |
| chr10_-_35321625 | 0.49 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr7_-_66864756 | 0.48 |
ENSDART00000184462
ENSDART00000189424 |
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr3_+_23654233 | 0.48 |
ENSDART00000078428
|
hoxb13a
|
homeobox B13a |
| chr6_+_23931236 | 0.48 |
ENSDART00000166079
|
gadd45ab
|
growth arrest and DNA-damage-inducible, alpha, b |
| chr15_+_9072821 | 0.47 |
ENSDART00000154463
|
si:dkey-202g17.3
|
si:dkey-202g17.3 |
| chr2_-_52455525 | 0.47 |
ENSDART00000160768
ENSDART00000002241 |
chico
|
chico |
| chr5_+_28857969 | 0.47 |
ENSDART00000149850
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr14_+_146857 | 0.46 |
ENSDART00000122521
|
CABZ01088229.1
|
|
| chr7_-_6357952 | 0.46 |
ENSDART00000173197
|
zgc:165555
|
zgc:165555 |
| chr16_+_11151699 | 0.46 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr17_+_8323348 | 0.45 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr5_+_19165949 | 0.44 |
ENSDART00000140710
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr24_-_11905911 | 0.44 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_-_19495814 | 0.44 |
ENSDART00000162248
|
CU571315.3
|
|
| chr3_-_2623176 | 0.43 |
ENSDART00000179792
ENSDART00000123512 |
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr7_+_7048245 | 0.42 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr5_+_28830388 | 0.42 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr13_+_2448251 | 0.41 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr16_-_42750295 | 0.41 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr19_+_43119698 | 0.41 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr1_-_8012476 | 0.40 |
ENSDART00000177051
|
CR855320.3
|
|
| chr13_-_37619159 | 0.40 |
ENSDART00000186348
|
zgc:152791
|
zgc:152791 |
| chr6_+_27339962 | 0.40 |
ENSDART00000193726
|
klhl30
|
kelch-like family member 30 |
| chr1_+_52560549 | 0.39 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr10_+_23520952 | 0.39 |
ENSDART00000125317
|
zmp:0000000937
|
zmp:0000000937 |
| chr9_-_39624173 | 0.39 |
ENSDART00000180106
ENSDART00000126766 |
erbb4b
|
erb-b2 receptor tyrosine kinase 4b |
| chr10_-_20065149 | 0.37 |
ENSDART00000182603
|
BX323022.1
|
|
| chr21_-_4755943 | 0.36 |
ENSDART00000137404
|
camsap1a
|
calmodulin regulated spectrin-associated protein 1a |
| chr3_+_58379450 | 0.36 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr5_+_28858345 | 0.35 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr23_-_31346319 | 0.35 |
ENSDART00000008401
ENSDART00000138106 |
phip
|
pleckstrin homology domain interacting protein |
| chr5_+_1911814 | 0.35 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr9_-_44983666 | 0.35 |
ENSDART00000149704
|
itgb2
|
integrin, beta 2 |
| chr15_-_25365319 | 0.34 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr22_-_906757 | 0.34 |
ENSDART00000193182
|
FP016205.1
|
|
| chr16_-_31451720 | 0.33 |
ENSDART00000146886
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr23_-_18057553 | 0.33 |
ENSDART00000173102
ENSDART00000058742 |
zgc:92287
|
zgc:92287 |
| chr18_+_8901846 | 0.33 |
ENSDART00000132109
ENSDART00000144247 |
si:dkey-5i3.5
|
si:dkey-5i3.5 |
| chr15_-_23647078 | 0.33 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr7_-_40578733 | 0.33 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr11_-_18604542 | 0.32 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr2_-_49997055 | 0.32 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr17_+_8292892 | 0.32 |
ENSDART00000125728
|
si:ch211-236p5.3
|
si:ch211-236p5.3 |
| chr4_-_4834347 | 0.32 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr18_+_14277003 | 0.32 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr24_-_28333029 | 0.32 |
ENSDART00000149015
ENSDART00000129174 |
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr21_-_2248239 | 0.31 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr6_-_39218609 | 0.31 |
ENSDART00000133305
|
os9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr15_-_727479 | 0.30 |
ENSDART00000157279
|
si:dkey-7i4.21
|
si:dkey-7i4.21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf21
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.8 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.4 | 1.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 4.0 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.3 | 1.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.3 | 1.0 | GO:0010657 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.0 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.3 | 1.8 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.2 | 0.7 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.2 | 1.7 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.9 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.2 | 1.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 2.1 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.2 | 1.0 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.7 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 1.0 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.1 | 2.9 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.5 | GO:0015859 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 1.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.7 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.0 | GO:0046247 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.8 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.4 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.1 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.1 | 0.6 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 0.9 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.3 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 0.4 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 9.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 0.9 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.7 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.1 | 0.3 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.1 | 0.8 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 3.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 | 1.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 1.6 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 4.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 1.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.2 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 2.0 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 8.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.5 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 0.4 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 2.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.0 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 1.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 1.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.9 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.6 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.3 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.7 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 1.8 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 2.7 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.3 | GO:0006090 | pyruvate metabolic process(GO:0006090) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.8 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 | 0.7 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.0 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 1.3 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.1 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.4 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 4.0 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 4.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.6 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.7 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.1 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 2.6 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.8 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.0 | 0.4 | GO:1990752 | microtubule minus-end(GO:0036449) microtubule end(GO:1990752) |
| 0.0 | 1.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 1.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.6 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.4 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.1 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.8 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.4 | GO:0042641 | actomyosin(GO:0042641) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.3 | 4.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.7 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.2 | 1.0 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.2 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 2.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.5 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 2.0 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 1.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 1.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 11.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.5 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.0 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 3.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.8 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 4.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.8 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 1.0 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.8 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.1 | 0.9 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.8 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 2.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.0 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.4 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.6 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.2 | GO:0032574 | 5'-3' RNA helicase activity(GO:0032574) |
| 0.1 | 0.9 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 5.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 2.0 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 3.3 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 2.6 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.7 | GO:0005501 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 3.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.6 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 1.5 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 2.1 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.5 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 2.5 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 2.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.3 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 2.0 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 0.6 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 1.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 1.0 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.3 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 0.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 3.5 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 1.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 1.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.3 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |