Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
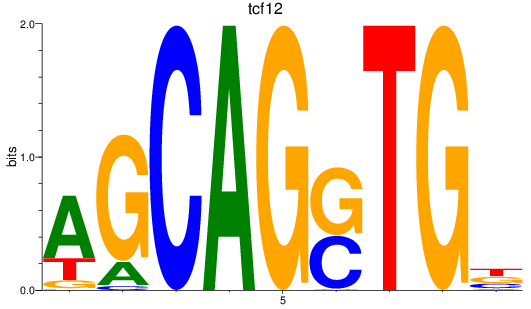
Results for tcf12
Z-value: 1.59
Transcription factors associated with tcf12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf12
|
ENSDARG00000004714 | transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf12 | dr11_v1_chr7_-_52709759_52709935 | 0.60 | 6.4e-03 | Click! |
Activity profile of tcf12 motif
Sorted Z-values of tcf12 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_38638809 | 4.81 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr11_-_101758 | 4.52 |
ENSDART00000173015
|
elmo2
|
engulfment and cell motility 2 |
| chr7_+_39444843 | 4.17 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr14_+_35901249 | 3.97 |
ENSDART00000105604
|
zgc:77938
|
zgc:77938 |
| chr23_-_7826849 | 3.57 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr19_+_233143 | 3.18 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr6_-_609880 | 2.95 |
ENSDART00000149248
ENSDART00000148867 ENSDART00000149414 ENSDART00000148552 ENSDART00000148391 |
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr23_+_28648864 | 2.94 |
ENSDART00000189096
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr16_+_3004422 | 2.93 |
ENSDART00000189969
|
CABZ01043952.1
|
|
| chr21_-_19018455 | 2.90 |
ENSDART00000080256
|
nefma
|
neurofilament, medium polypeptide a |
| chr5_+_42467867 | 2.84 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr9_-_22892838 | 2.83 |
ENSDART00000143888
|
neb
|
nebulin |
| chr5_-_38384289 | 2.77 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr17_-_37395460 | 2.69 |
ENSDART00000148160
ENSDART00000075975 |
crip1
|
cysteine-rich protein 1 |
| chr16_+_4055331 | 2.68 |
ENSDART00000128978
|
CR391998.1
|
|
| chr11_+_20371179 | 2.63 |
ENSDART00000104022
ENSDART00000164982 |
cdh4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr9_-_44948488 | 2.61 |
ENSDART00000059228
|
vil1
|
villin 1 |
| chr7_-_38638276 | 2.57 |
ENSDART00000074463
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr24_-_40009446 | 2.56 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr2_-_22688651 | 2.55 |
ENSDART00000013863
|
agxtb
|
alanine-glyoxylate aminotransferase b |
| chr11_+_7324704 | 2.55 |
ENSDART00000031937
|
diras1a
|
DIRAS family, GTP-binding RAS-like 1a |
| chr3_-_28075756 | 2.47 |
ENSDART00000122037
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr13_-_27916439 | 2.46 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr20_-_52902693 | 2.44 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr21_+_11468642 | 2.43 |
ENSDART00000041869
|
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr18_-_5781922 | 2.40 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr13_+_30054996 | 2.39 |
ENSDART00000110061
ENSDART00000186045 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr11_+_23993298 | 2.39 |
ENSDART00000186757
ENSDART00000172459 |
chia.1
|
chitinase, acidic.1 |
| chr8_-_25247284 | 2.32 |
ENSDART00000132697
|
gnat2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 |
| chr15_-_37829160 | 2.31 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr25_-_169291 | 2.30 |
ENSDART00000128344
|
lipcb
|
lipase, hepatic b |
| chr18_-_14274803 | 2.29 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr3_-_27880229 | 2.26 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr3_+_32526799 | 2.25 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr25_-_3549321 | 2.25 |
ENSDART00000181214
ENSDART00000160600 |
hdhd5
|
haloacid dehalogenase like hydrolase domain containing 5 |
| chr13_+_29462249 | 2.24 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr6_-_16667886 | 2.23 |
ENSDART00000180854
ENSDART00000190116 |
unc80
|
unc-80 homolog (C. elegans) |
| chr18_-_5692292 | 2.22 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr18_-_8312848 | 2.22 |
ENSDART00000092033
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr1_-_38816685 | 2.19 |
ENSDART00000075230
|
asb5b
|
ankyrin repeat and SOCS box containing 5b |
| chr3_-_36115339 | 2.18 |
ENSDART00000187406
ENSDART00000123505 ENSDART00000151775 |
rab11fip4a
|
RAB11 family interacting protein 4 (class II) a |
| chr11_-_669270 | 2.16 |
ENSDART00000172834
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr8_+_21376290 | 2.15 |
ENSDART00000136765
|
ela2
|
elastase 2 |
| chr12_-_4756478 | 2.13 |
ENSDART00000152181
|
mapta
|
microtubule-associated protein tau a |
| chr5_-_38342992 | 2.12 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr11_+_1845787 | 2.12 |
ENSDART00000173062
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr24_-_31452875 | 2.11 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr24_-_26283359 | 2.10 |
ENSDART00000128618
|
and1
|
actinodin1 |
| chr4_-_14328997 | 2.07 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr16_-_44127307 | 2.07 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr13_+_30055171 | 2.04 |
ENSDART00000143581
ENSDART00000132027 |
spock2
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr7_+_10610791 | 2.02 |
ENSDART00000166064
|
fah
|
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
| chr13_-_37200120 | 2.00 |
ENSDART00000135001
|
si:dkeyp-77c8.3
|
si:dkeyp-77c8.3 |
| chr22_-_14128716 | 1.99 |
ENSDART00000140323
|
si:ch211-246m6.4
|
si:ch211-246m6.4 |
| chr1_-_21409877 | 1.99 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr13_-_49846301 | 1.98 |
ENSDART00000182293
|
GNG4
|
G protein subunit gamma 4 |
| chr14_-_4556896 | 1.96 |
ENSDART00000044678
ENSDART00000192863 |
GABRA2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr3_-_30861177 | 1.92 |
ENSDART00000154811
|
shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr17_-_722218 | 1.92 |
ENSDART00000160385
|
SLC25A29
|
solute carrier family 25 member 29 |
| chr16_-_13612650 | 1.90 |
ENSDART00000080372
|
dbpb
|
D site albumin promoter binding protein b |
| chr21_+_11468934 | 1.89 |
ENSDART00000126045
ENSDART00000129744 ENSDART00000102368 |
grin1a
|
glutamate receptor, ionotropic, N-methyl D-aspartate 1a |
| chr11_+_30161168 | 1.88 |
ENSDART00000157385
|
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr23_-_26535875 | 1.87 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr16_+_43152727 | 1.83 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr4_+_3482312 | 1.83 |
ENSDART00000109044
|
grm8a
|
glutamate receptor, metabotropic 8a |
| chr2_+_15612755 | 1.82 |
ENSDART00000003035
|
amy2a
|
amylase, alpha 2A (pancreatic) |
| chr1_-_25911292 | 1.82 |
ENSDART00000145012
|
usp53b
|
ubiquitin specific peptidase 53b |
| chr23_-_1583193 | 1.81 |
ENSDART00000143841
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr20_-_53624645 | 1.80 |
ENSDART00000083427
ENSDART00000152920 |
slc25a29
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine carrier), member 29 |
| chr13_+_22264914 | 1.79 |
ENSDART00000060576
|
myoz1a
|
myozenin 1a |
| chr11_-_4220167 | 1.79 |
ENSDART00000185406
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr8_-_23416362 | 1.79 |
ENSDART00000063005
|
gpr173
|
G protein-coupled receptor 173 |
| chr6_+_55006686 | 1.78 |
ENSDART00000081703
|
nav1b
|
neuron navigator 1b |
| chr24_-_38384432 | 1.77 |
ENSDART00000140739
|
lrrc4bb
|
leucine rich repeat containing 4Bb |
| chr5_-_13766651 | 1.76 |
ENSDART00000134064
|
mxd1
|
MAX dimerization protein 1 |
| chr7_+_22616212 | 1.75 |
ENSDART00000052844
|
cldn7a
|
claudin 7a |
| chr21_-_27123139 | 1.73 |
ENSDART00000077478
|
slc8a4a
|
solute carrier family 8 (sodium/calcium exchanger), member 4a |
| chr4_+_5531583 | 1.73 |
ENSDART00000137500
ENSDART00000042080 |
si:dkey-14d8.6
|
si:dkey-14d8.6 |
| chr14_+_35748385 | 1.72 |
ENSDART00000064617
ENSDART00000074671 ENSDART00000172803 |
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr16_+_46148990 | 1.71 |
ENSDART00000083919
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr18_-_40684756 | 1.70 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr14_+_45925810 | 1.69 |
ENSDART00000189543
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr1_+_14432434 | 1.69 |
ENSDART00000024328
|
slc34a2a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 2a |
| chr2_+_30878864 | 1.67 |
ENSDART00000009326
|
oprk1
|
opioid receptor, kappa 1 |
| chr13_+_1575276 | 1.66 |
ENSDART00000165987
|
DST
|
dystonin |
| chr23_-_16692312 | 1.66 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr24_-_14711597 | 1.66 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr1_-_50838160 | 1.66 |
ENSDART00000163939
ENSDART00000165111 |
zgc:154142
|
zgc:154142 |
| chr9_-_43375205 | 1.65 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr23_-_3674443 | 1.65 |
ENSDART00000134830
ENSDART00000057422 |
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr22_-_21897203 | 1.65 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr15_+_19915345 | 1.65 |
ENSDART00000114267
|
map6b
|
microtubule-associated protein 6b |
| chr13_-_9318891 | 1.64 |
ENSDART00000137364
|
si:dkey-33c12.3
|
si:dkey-33c12.3 |
| chr25_-_19433244 | 1.62 |
ENSDART00000154778
|
map1ab
|
microtubule-associated protein 1Ab |
| chr10_-_41450367 | 1.62 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr23_+_29908348 | 1.62 |
ENSDART00000186622
ENSDART00000150174 |
si:ch73-236e11.2
|
si:ch73-236e11.2 |
| chr15_+_19915772 | 1.61 |
ENSDART00000188911
|
map6b
|
microtubule-associated protein 6b |
| chr8_+_26141680 | 1.61 |
ENSDART00000078334
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr25_-_20258508 | 1.60 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr16_-_36798783 | 1.60 |
ENSDART00000145697
|
calb1
|
calbindin 1 |
| chr17_-_5610514 | 1.60 |
ENSDART00000004043
|
enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr7_+_31891110 | 1.59 |
ENSDART00000173883
|
mybpc3
|
myosin binding protein C, cardiac |
| chr21_-_5007109 | 1.59 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr14_+_35748206 | 1.59 |
ENSDART00000177391
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr6_+_55277419 | 1.59 |
ENSDART00000083670
|
CABZ01041604.1
|
|
| chr9_-_49531762 | 1.58 |
ENSDART00000121875
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr5_+_3501859 | 1.58 |
ENSDART00000080486
|
ywhag1
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 1 |
| chr13_+_33703784 | 1.58 |
ENSDART00000173361
|
macrod2
|
MACRO domain containing 2 |
| chr5_-_24238733 | 1.57 |
ENSDART00000138170
|
plscr3a
|
phospholipid scramblase 3a |
| chr12_+_45676667 | 1.57 |
ENSDART00000016553
|
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr7_+_22767678 | 1.57 |
ENSDART00000137203
|
ponzr6
|
plac8 onzin related protein 6 |
| chr6_+_54888493 | 1.57 |
ENSDART00000113331
|
nav1b
|
neuron navigator 1b |
| chr1_+_8662530 | 1.57 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr25_-_35542739 | 1.56 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr20_-_18731268 | 1.56 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr25_-_207214 | 1.56 |
ENSDART00000193448
|
FP236318.3
|
|
| chr17_-_16069905 | 1.55 |
ENSDART00000110383
|
map7a
|
microtubule-associated protein 7a |
| chr1_-_54158902 | 1.55 |
ENSDART00000193267
|
CABZ01067151.1
|
|
| chr23_+_6795531 | 1.54 |
ENSDART00000092131
|
si:ch211-117c9.5
|
si:ch211-117c9.5 |
| chr19_+_43669122 | 1.54 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr15_+_28482862 | 1.54 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr13_+_27314795 | 1.53 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr15_-_47937204 | 1.52 |
ENSDART00000154705
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr8_-_46700278 | 1.52 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr21_-_43606502 | 1.51 |
ENSDART00000151030
|
si:ch73-362m14.4
|
si:ch73-362m14.4 |
| chr6_+_29305190 | 1.51 |
ENSDART00000078647
|
si:ch211-201h21.5
|
si:ch211-201h21.5 |
| chr20_-_37629084 | 1.51 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr16_-_25563508 | 1.51 |
ENSDART00000189642
|
atxn1b
|
ataxin 1b |
| chr15_+_47386939 | 1.50 |
ENSDART00000128224
|
FO904873.1
|
|
| chr5_+_36781732 | 1.49 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr25_-_8030425 | 1.49 |
ENSDART00000014964
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr16_-_17207754 | 1.49 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr3_-_32169754 | 1.49 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr3_-_3209432 | 1.49 |
ENSDART00000140635
|
si:ch211-229i14.2
|
si:ch211-229i14.2 |
| chr17_+_15845765 | 1.48 |
ENSDART00000130881
ENSDART00000074936 |
gabrr2a
|
gamma-aminobutyric acid (GABA) A receptor, rho 2a |
| chr11_-_36963988 | 1.48 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr5_+_67812062 | 1.48 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr3_+_32526263 | 1.47 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr2_+_38881165 | 1.47 |
ENSDART00000141850
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr20_+_27020201 | 1.47 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr17_-_19828505 | 1.46 |
ENSDART00000154745
|
fmn2a
|
formin 2a |
| chr5_+_23118470 | 1.46 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr12_-_7710563 | 1.45 |
ENSDART00000187382
|
CABZ01050062.1
|
|
| chr13_+_2448251 | 1.45 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr22_+_32120156 | 1.45 |
ENSDART00000149666
|
dock3
|
dedicator of cytokinesis 3 |
| chr22_-_968484 | 1.45 |
ENSDART00000105895
|
cacna1sa
|
calcium channel, voltage-dependent, L type, alpha 1S subunit, a |
| chr19_-_13774502 | 1.44 |
ENSDART00000159711
|
epb41a
|
erythrocyte membrane protein band 4.1a |
| chr23_-_18668836 | 1.44 |
ENSDART00000138792
ENSDART00000051182 |
arhgap4b
|
Rho GTPase activating protein 4b |
| chr21_-_43952958 | 1.43 |
ENSDART00000039571
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_-_1291012 | 1.42 |
ENSDART00000158390
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr9_-_3653259 | 1.42 |
ENSDART00000140425
ENSDART00000025332 |
gad1a
|
glutamate decarboxylase 1a |
| chr11_+_30000814 | 1.42 |
ENSDART00000191011
ENSDART00000189770 |
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr10_-_43964028 | 1.42 |
ENSDART00000009134
ENSDART00000133450 |
sept5b
|
septin 5b |
| chr22_-_57177 | 1.42 |
ENSDART00000163959
|
CABZ01085139.1
|
|
| chr23_+_40452157 | 1.41 |
ENSDART00000113106
ENSDART00000140136 |
soga3b
|
SOGA family member 3b |
| chr22_+_5106751 | 1.41 |
ENSDART00000138967
|
atcaya
|
ataxia, cerebellar, Cayman type a |
| chr7_+_31145386 | 1.40 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr11_+_6819050 | 1.39 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr6_-_29305132 | 1.38 |
ENSDART00000132456
|
bivm
|
basic, immunoglobulin-like variable motif containing |
| chr20_+_41549200 | 1.38 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr23_+_21067684 | 1.38 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr21_+_30673183 | 1.37 |
ENSDART00000144652
|
clcn5a
|
chloride channel, voltage-sensitive 5a |
| chr2_-_42393590 | 1.37 |
ENSDART00000135529
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr18_+_402048 | 1.37 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr25_-_19655820 | 1.36 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr5_+_2815021 | 1.36 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr8_-_53198154 | 1.36 |
ENSDART00000083416
|
gabrd
|
gamma-aminobutyric acid (GABA) A receptor, delta |
| chr5_+_20148671 | 1.36 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr21_-_131236 | 1.35 |
ENSDART00000160005
|
si:ch1073-398f15.1
|
si:ch1073-398f15.1 |
| chr8_+_24861264 | 1.35 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr21_+_27382893 | 1.35 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr25_+_37285737 | 1.35 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr18_-_15771551 | 1.34 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr2_-_8111346 | 1.34 |
ENSDART00000123103
|
pls1
|
plastin 1 (I isoform) |
| chr5_-_22952156 | 1.34 |
ENSDART00000111146
|
si:ch211-26b3.4
|
si:ch211-26b3.4 |
| chr4_+_23223881 | 1.34 |
ENSDART00000133056
ENSDART00000089126 |
trhde.1
|
thyrotropin releasing hormone degrading enzyme, tandem duplicate 1 |
| chr15_-_7598294 | 1.34 |
ENSDART00000165898
|
gbe1b
|
glucan (1,4-alpha-), branching enzyme 1b |
| chr15_+_30158652 | 1.33 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr3_+_42923275 | 1.33 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr18_+_15271993 | 1.33 |
ENSDART00000099777
|
si:dkey-103i16.6
|
si:dkey-103i16.6 |
| chr4_+_12617108 | 1.33 |
ENSDART00000134362
ENSDART00000112860 |
lmo3
|
LIM domain only 3 |
| chr10_-_39153959 | 1.32 |
ENSDART00000150193
ENSDART00000111362 |
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr4_-_9549693 | 1.32 |
ENSDART00000160242
|
FQ377934.1
|
|
| chr13_-_11048063 | 1.32 |
ENSDART00000185831
|
cep170aa
|
centrosomal protein 170Aa |
| chr2_-_37312927 | 1.32 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr11_-_43226255 | 1.32 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr13_+_25449681 | 1.32 |
ENSDART00000101328
|
atoh7
|
atonal bHLH transcription factor 7 |
| chr4_-_8043839 | 1.32 |
ENSDART00000190047
ENSDART00000057567 |
si:ch211-240l19.5
|
si:ch211-240l19.5 |
| chr8_-_10048059 | 1.31 |
ENSDART00000138411
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr1_-_21909608 | 1.31 |
ENSDART00000139937
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr2_+_8261109 | 1.31 |
ENSDART00000024677
|
dia1a
|
deleted in autism 1a |
| chr7_+_26224211 | 1.31 |
ENSDART00000173999
|
vgf
|
VGF nerve growth factor inducible |
| chr5_-_41831646 | 1.31 |
ENSDART00000134326
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr21_-_45588720 | 1.31 |
ENSDART00000186642
ENSDART00000189531 |
LO018363.2
|
|
| chr12_-_48006835 | 1.31 |
ENSDART00000108989
|
adamts14
|
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
| chr9_-_18568927 | 1.30 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr5_+_6796291 | 1.30 |
ENSDART00000166868
ENSDART00000165308 |
me2
|
malic enzyme 2, NAD(+)-dependent, mitochondrial |
| chr21_-_24865454 | 1.30 |
ENSDART00000142907
|
igsf9bb
|
immunoglobulin superfamily, member 9Bb |
| chr10_-_22831611 | 1.30 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr5_-_13076779 | 1.29 |
ENSDART00000192826
|
ypel1
|
yippee-like 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf12
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.9 | 3.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.8 | 3.9 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.7 | 2.6 | GO:0042478 | regulation of eye photoreceptor cell development(GO:0042478) |
| 0.7 | 2.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.6 | 4.0 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.6 | 3.4 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.6 | 1.7 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.5 | 2.6 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.5 | 2.5 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.5 | 2.5 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.5 | 2.9 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.5 | 1.9 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.5 | 2.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 1.4 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.5 | 2.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.5 | 1.8 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.5 | 1.4 | GO:0015824 | proline transport(GO:0015824) |
| 0.4 | 1.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.4 | 1.7 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.4 | 1.7 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 1.6 | GO:0099509 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 1.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.4 | 1.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.4 | 1.8 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.4 | 3.9 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 2.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.3 | 1.0 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.3 | 1.0 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.3 | 1.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.3 | 1.5 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 1.5 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.3 | 1.2 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.3 | 1.7 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.3 | 5.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.3 | 2.0 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.3 | 0.8 | GO:0000256 | allantoin catabolic process(GO:0000256) |
| 0.3 | 6.4 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 0.8 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.3 | 1.3 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.3 | 1.8 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.3 | 0.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.3 | 3.0 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.3 | 0.8 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 0.7 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 | 1.5 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.2 | 0.7 | GO:2000402 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.2 | 1.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 1.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.2 | 1.2 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.2 | 1.4 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.2 | 0.5 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.2 | 0.7 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.2 | 1.6 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.2 | 2.8 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.2 | 2.4 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 2.4 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 2.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.2 | 1.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 0.8 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.2 | 1.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 1.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 1.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 0.8 | GO:0015677 | copper ion import(GO:0015677) |
| 0.2 | 0.9 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.2 | 0.6 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.2 | 4.5 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.2 | 0.9 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.2 | 0.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 1.4 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 0.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.2 | 0.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 1.8 | GO:0042044 | fluid transport(GO:0042044) |
| 0.2 | 2.0 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.2 | 0.6 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.2 | 2.9 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.2 | 1.1 | GO:1902902 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 2.4 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.1 | 1.3 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 0.4 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 0.7 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.7 | GO:0006921 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 2.0 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 0.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.7 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.6 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 0.8 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 6.0 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.5 | GO:0002698 | negative regulation of immune effector process(GO:0002698) |
| 0.1 | 1.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 1.7 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 3.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 0.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.0 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.6 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 1.5 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.3 | GO:0060945 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.8 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 0.8 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 1.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 1.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 2.1 | GO:0034315 | regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 0.7 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.3 | GO:0032757 | interleukin-8 production(GO:0032637) regulation of interleukin-8 production(GO:0032677) positive regulation of interleukin-8 production(GO:0032757) |
| 0.1 | 1.9 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 1.1 | GO:0061620 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.1 | 0.7 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 6.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.8 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.3 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 1.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.1 | 4.2 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 0.8 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 6.2 | GO:0032273 | positive regulation of protein polymerization(GO:0032273) |
| 0.1 | 0.3 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 1.0 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.1 | 0.5 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.4 | GO:0008207 | C21-steroid hormone metabolic process(GO:0008207) |
| 0.1 | 1.4 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.1 | 1.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 0.4 | GO:0014896 | muscle hypertrophy(GO:0014896) |
| 0.1 | 1.8 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.5 | GO:0061440 | renal system vasculature development(GO:0061437) kidney vasculature development(GO:0061440) glomerulus vasculature development(GO:0072012) |
| 0.1 | 1.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 1.1 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 0.3 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.6 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.1 | 5.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 4.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.0 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 3.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.8 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.7 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.1 | 1.8 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 1.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.1 | 0.8 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 2.3 | GO:0050433 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) catecholamine secretion(GO:0050432) regulation of catecholamine secretion(GO:0050433) |
| 0.1 | 0.5 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.3 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 2.4 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 1.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.6 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.8 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 1.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 3.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 0.8 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.0 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.6 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 1.6 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 1.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.1 | 0.9 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) dendritic spine organization(GO:0097061) |
| 0.1 | 1.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 3.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 0.9 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.6 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.1 | 0.7 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.1 | 2.1 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.0 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 1.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.5 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 4.0 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.1 | 1.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.7 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 1.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.4 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.1 | 1.6 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.2 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.1 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.4 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.5 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 1.1 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 1.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.4 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 0.3 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.3 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.3 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.0 | 0.7 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.2 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 1.8 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.3 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 1.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.0 | GO:0071230 | response to amino acid(GO:0043200) cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.7 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 3.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 4.6 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 1.2 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.6 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 2.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.3 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 1.4 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.3 | GO:1900026 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.7 | GO:0010906 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.3 | GO:0000423 | macromitophagy(GO:0000423) C-terminal protein lipidation(GO:0006501) lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) single-organism membrane invagination(GO:1902534) |
| 0.0 | 4.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.6 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.7 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 1.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.1 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.0 | 0.2 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.5 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 3.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 1.1 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 3.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 2.2 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 2.3 | GO:0030041 | actin filament polymerization(GO:0030041) |
| 0.0 | 0.5 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.5 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 4.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.1 | GO:0071678 | anterior commissure morphogenesis(GO:0021960) olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 1.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.4 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.0 | 2.4 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.9 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.1 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.3 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.0 | 0.4 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.7 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:1903513 | endoplasmic reticulum to cytosol transport(GO:1903513) |
| 0.0 | 0.2 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.4 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.2 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.2 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0045923 | positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.0 | 1.7 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.0 | 0.8 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.5 | GO:0043491 | protein kinase B signaling(GO:0043491) |
| 0.0 | 0.6 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.1 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.5 | GO:0097202 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.0 | 0.2 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 0.1 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.0 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.1 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.3 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.8 | GO:0007599 | hemostasis(GO:0007599) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.9 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 1.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.5 | GO:0097696 | JAK-STAT cascade(GO:0007259) STAT cascade(GO:0097696) |
| 0.0 | 1.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 1.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.0 | GO:0036089 | cytokinetic process(GO:0032506) cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0032412 | regulation of ion transmembrane transporter activity(GO:0032412) |
| 0.0 | 0.2 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.3 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0031529 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.0 | 0.7 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.6 | 2.9 | GO:0005883 | neurofilament(GO:0005883) |
| 0.5 | 2.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 3.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.5 | 2.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 2.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 5.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 1.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.3 | 3.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 3.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.3 | 2.7 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.3 | 1.0 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 4.1 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 4.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.2 | 1.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.2 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.0 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 1.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 5.5 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 6.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.9 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.9 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 3.1 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 1.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.8 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.5 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 5.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 2.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.9 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 3.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.5 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.5 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 1.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.7 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 1.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 4.5 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 6.8 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.1 | 0.9 | GO:0043679 | axon terminus(GO:0043679) |
| 0.1 | 1.3 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 4.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.3 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 11.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.1 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 3.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.4 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 1.4 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 4.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 5.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 3.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 2.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 4.0 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.4 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.8 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.9 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 3.3 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.6 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.2 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 1.9 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.3 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.8 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 1.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 2.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 4.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 4.2 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 3.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.0 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.0 | 0.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 1.6 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.8 | 4.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.8 | 2.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.6 | 3.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.6 | 2.9 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.6 | 5.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.5 | 2.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.5 | 1.4 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.5 | 1.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.5 | 1.8 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.4 | 4.8 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.4 | 2.0 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.4 | 3.9 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 5.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.3 | 1.0 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.3 | 1.3 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.3 | 2.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 1.3 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.3 | 1.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 1.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.3 | 0.9 | GO:0050135 | NAD(P)+ nucleosidase activity(GO:0050135) |
| 0.3 | 3.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 1.5 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.3 | 2.9 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.3 | 1.7 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.3 | 2.7 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.3 | 1.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 2.3 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.3 | 5.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 2.4 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.2 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 2.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.9 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 1.0 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.2 | 1.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 0.8 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.2 | 2.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.2 | 5.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 0.9 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 0.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.2 | 1.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 4.0 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.2 | 1.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.2 | 3.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 1.5 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.8 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.2 | 1.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 1.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.2 | 5.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 1.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 0.6 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 0.5 | GO:0047173 | phosphatidylcholine-retinol O-acyltransferase activity(GO:0047173) |
| 0.2 | 2.8 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.2 | 0.9 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 2.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 0.6 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.4 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 1.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.4 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.4 | GO:0004777 | succinate-semialdehyde dehydrogenase (NAD+) activity(GO:0004777) succinate-semialdehyde dehydrogenase [NAD(P)+] activity(GO:0009013) |
| 0.1 | 0.6 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 1.5 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.6 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 1.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 2.1 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 1.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.8 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 3.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.3 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.1 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.8 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 2.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 3.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 1.6 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 2.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 2.2 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.1 | 5.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 2.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 3.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 0.4 | GO:0070699 | adrenergic receptor binding(GO:0031690) beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.9 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 1.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 5.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.8 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.5 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.8 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.8 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 2.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 2.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.2 | GO:0008443 | 6-phosphofructo-2-kinase activity(GO:0003873) phosphofructokinase activity(GO:0008443) |
| 0.1 | 0.8 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 12.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.4 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 4.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 3.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.8 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 3.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 2.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 2.0 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.6 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.8 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 0.6 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 3.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.5 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.6 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 1.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 2.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.9 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 1.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.3 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.1 | 1.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 5.0 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.2 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 1.7 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.4 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.3 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.2 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.1 | 0.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.9 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.7 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.7 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.5 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 1.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 1.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.9 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0035255 | GKAP/Homer scaffold activity(GO:0030160) ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.3 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.7 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 12.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.0 | 10.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 1.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.6 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0008169 | C-methyltransferase activity(GO:0008169) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 1.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 1.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.2 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 3.0 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.2 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 7.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.2 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 1.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 2.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 4.9 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 4.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0005501 | retinoid binding(GO:0005501) isoprenoid binding(GO:0019840) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 2.5 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.2 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 3.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 4.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 1.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.2 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.1 | 2.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.1 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.3 | 0.6 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.3 | 4.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 2.6 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 0.6 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 4.3 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.2 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 4.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 1.0 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 2.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 3.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 1.0 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 2.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.5 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 1.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.1 | 0.8 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.2 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.1 | 2.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 2.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 3.5 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.7 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 1.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 1.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 1.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 2.3 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.4 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.2 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |