Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
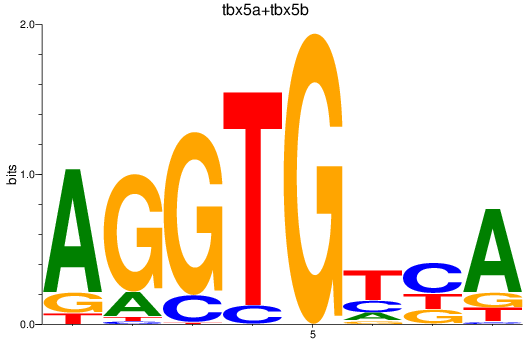
Results for tbx5a+tbx5b
Z-value: 0.60
Transcription factors associated with tbx5a+tbx5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx5a
|
ENSDARG00000024894 | T-box transcription factor 5a |
|
tbx5b
|
ENSDARG00000092060 | T-box transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx5a | dr11_v1_chr5_-_72289648_72289648 | 0.78 | 7.8e-05 | Click! |
| tbx5b | dr11_v1_chr5_+_23151169_23151169 | 0.65 | 2.7e-03 | Click! |
Activity profile of tbx5a+tbx5b motif
Sorted Z-values of tbx5a+tbx5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_293036 | 1.51 |
ENSDART00000171629
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr5_+_26075230 | 1.38 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr21_+_25226558 | 1.32 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr7_-_38638809 | 1.16 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr23_-_30785382 | 0.99 |
ENSDART00000136156
ENSDART00000131285 |
myt1a
|
myelin transcription factor 1a |
| chr10_-_2522588 | 0.86 |
ENSDART00000081926
|
CU856539.1
|
|
| chr12_-_20362041 | 0.83 |
ENSDART00000184145
ENSDART00000105952 |
aqp8a.2
|
aquaporin 8a, tandem duplicate 2 |
| chr18_+_15841449 | 0.81 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr13_+_47821524 | 0.81 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr7_+_73780936 | 0.81 |
ENSDART00000092691
|
pea15
|
proliferation and apoptosis adaptor protein 15 |
| chr13_-_3516473 | 0.78 |
ENSDART00000146240
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr9_-_44948488 | 0.78 |
ENSDART00000059228
|
vil1
|
villin 1 |
| chr21_-_10488379 | 0.78 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr12_-_31457301 | 0.75 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr23_+_44263986 | 0.75 |
ENSDART00000149194
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr15_-_25365319 | 0.71 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr16_+_3982590 | 0.71 |
ENSDART00000149295
|
zc3h12a
|
zinc finger CCCH-type containing 12A |
| chr13_+_6342410 | 0.68 |
ENSDART00000065228
|
csmd1a
|
CUB and Sushi multiple domains 1a |
| chr22_-_12160283 | 0.65 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr7_+_36539124 | 0.64 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr13_-_31888487 | 0.63 |
ENSDART00000145938
|
syt14a
|
synaptotagmin XIVa |
| chr3_+_19621034 | 0.63 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr25_-_4482449 | 0.62 |
ENSDART00000056278
ENSDART00000149425 |
slc25a22a
|
solute carrier family 25 member 22a |
| chr17_+_8212477 | 0.61 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr3_+_12732382 | 0.61 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr13_-_290377 | 0.61 |
ENSDART00000134963
|
chs1
|
chitin synthase 1 |
| chr23_-_12158685 | 0.60 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr3_-_53508580 | 0.58 |
ENSDART00000073978
|
zgc:171711
|
zgc:171711 |
| chr24_-_38079261 | 0.58 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr11_+_1628538 | 0.57 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr23_+_36601984 | 0.57 |
ENSDART00000128598
|
igfbp6b
|
insulin-like growth factor binding protein 6b |
| chr16_-_21668082 | 0.55 |
ENSDART00000088513
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chr5_+_13472234 | 0.55 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr23_+_21067684 | 0.54 |
ENSDART00000132066
|
kcnab2b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 b |
| chr16_+_26774182 | 0.54 |
ENSDART00000042895
|
cdh17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr3_+_49074008 | 0.53 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr1_-_9231952 | 0.53 |
ENSDART00000166515
|
si:dkeyp-57d7.4
|
si:dkeyp-57d7.4 |
| chr3_+_17456428 | 0.52 |
ENSDART00000090676
ENSDART00000182082 |
si:ch211-210g13.5
|
si:ch211-210g13.5 |
| chr17_-_15611744 | 0.52 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr12_+_35119762 | 0.52 |
ENSDART00000085774
|
si:ch73-127m5.1
|
si:ch73-127m5.1 |
| chr18_-_14274803 | 0.52 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr15_-_551177 | 0.51 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr2_+_32835679 | 0.51 |
ENSDART00000132767
|
pxdc1a
|
PX domain containing 1a |
| chr8_+_30456161 | 0.51 |
ENSDART00000085894
|
pgm5
|
phosphoglucomutase 5 |
| chr3_+_60607241 | 0.51 |
ENSDART00000167512
|
mfsd11
|
major facilitator superfamily domain containing 11 |
| chr3_+_41726360 | 0.51 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr21_-_35325466 | 0.51 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr15_+_19915772 | 0.50 |
ENSDART00000188911
|
map6b
|
microtubule-associated protein 6b |
| chr24_-_6501211 | 0.50 |
ENSDART00000186241
ENSDART00000109040 ENSDART00000136154 |
gpr158a
|
G protein-coupled receptor 158a |
| chr18_+_13077800 | 0.50 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr8_+_46418996 | 0.50 |
ENSDART00000144285
|
si:ch211-196g2.4
|
si:ch211-196g2.4 |
| chr2_+_45344070 | 0.50 |
ENSDART00000147245
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr3_-_39171968 | 0.49 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr8_+_14778292 | 0.49 |
ENSDART00000089971
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr16_-_2558653 | 0.48 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr14_+_45406299 | 0.48 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr3_-_24909245 | 0.48 |
ENSDART00000178321
|
SHISA8
|
shisa family member 8 |
| chr24_-_11127493 | 0.48 |
ENSDART00000144066
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr16_-_17207754 | 0.48 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr22_-_29242347 | 0.47 |
ENSDART00000040761
|
pvalb7
|
parvalbumin 7 |
| chr5_+_9360394 | 0.47 |
ENSDART00000124642
|
FP236810.2
|
|
| chr12_-_36045283 | 0.47 |
ENSDART00000160646
|
gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr8_+_2478862 | 0.47 |
ENSDART00000131739
|
si:dkeyp-51b9.3
|
si:dkeyp-51b9.3 |
| chr25_-_17528994 | 0.46 |
ENSDART00000061712
|
si:dkey-44k1.5
|
si:dkey-44k1.5 |
| chr23_+_27912079 | 0.46 |
ENSDART00000171859
|
BX539336.1
|
|
| chr6_-_51665454 | 0.45 |
ENSDART00000028607
|
chd6
|
chromodomain helicase DNA binding protein 6 |
| chr24_-_2312868 | 0.45 |
ENSDART00000140125
ENSDART00000138432 |
cul2
|
cullin 2 |
| chr18_+_33580391 | 0.45 |
ENSDART00000189300
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr12_+_19976400 | 0.44 |
ENSDART00000153177
|
mkl2a
|
MKL/myocardin-like 2a |
| chr3_-_12352793 | 0.44 |
ENSDART00000165013
|
pam16
|
presequence translocase associated motor 16 |
| chr23_-_637347 | 0.43 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr25_+_32525131 | 0.42 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr3_+_12710350 | 0.41 |
ENSDART00000157959
|
cyp2k18
|
cytochrome P450, family 2, subfamily K, polypeptide 18 |
| chr2_-_2642476 | 0.41 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr9_-_30165621 | 0.41 |
ENSDART00000089543
|
abi3bpa
|
ABI family, member 3 (NESH) binding protein a |
| chr8_+_52637507 | 0.41 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr7_-_51476276 | 0.41 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr23_-_270847 | 0.41 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr10_+_34426571 | 0.41 |
ENSDART00000144529
|
nbeaa
|
neurobeachin a |
| chr19_-_6983002 | 0.41 |
ENSDART00000104891
|
znf384l
|
zinc finger protein 384 like |
| chr12_+_16168342 | 0.41 |
ENSDART00000079326
ENSDART00000170024 |
lrp2b
|
low density lipoprotein receptor-related protein 2b |
| chr9_+_52492639 | 0.41 |
ENSDART00000078939
|
march4
|
membrane-associated ring finger (C3HC4) 4 |
| chr4_+_16768961 | 0.40 |
ENSDART00000143926
|
BX649500.1
|
|
| chr23_-_45504991 | 0.40 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr7_+_65309738 | 0.40 |
ENSDART00000169228
|
vat1l
|
vesicle amine transport 1-like |
| chr21_+_25187210 | 0.40 |
ENSDART00000101147
ENSDART00000167528 |
si:dkey-183i3.5
|
si:dkey-183i3.5 |
| chr17_+_36588281 | 0.40 |
ENSDART00000076115
|
htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr5_-_69482891 | 0.39 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr20_-_8419057 | 0.39 |
ENSDART00000145841
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr1_-_6085750 | 0.39 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr15_+_38887467 | 0.39 |
ENSDART00000185222
|
robo2
|
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr1_-_22512063 | 0.39 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr14_-_17121676 | 0.38 |
ENSDART00000170154
ENSDART00000060479 |
smtnl1
|
smoothelin-like 1 |
| chr22_+_26813157 | 0.38 |
ENSDART00000140261
|
slx4
|
SLX4 structure-specific endonuclease subunit homolog (S. cerevisiae) |
| chr3_+_16762483 | 0.38 |
ENSDART00000132732
|
tmem86b
|
transmembrane protein 86B |
| chr23_+_19790962 | 0.38 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr17_+_3124129 | 0.38 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr15_-_43164591 | 0.38 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr8_+_7144066 | 0.38 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr5_+_4436405 | 0.38 |
ENSDART00000167969
|
CABZ01079241.1
|
|
| chr24_+_8904135 | 0.38 |
ENSDART00000066782
|
gnal
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr10_+_6907715 | 0.38 |
ENSDART00000041068
|
slc38a9
|
solute carrier family 38, member 9 |
| chr9_+_38088331 | 0.38 |
ENSDART00000123749
|
cacnb4a
|
calcium channel, voltage-dependent, beta 4a subunit |
| chr10_+_23548674 | 0.37 |
ENSDART00000079686
|
zmp:0000001103
|
zmp:0000001103 |
| chr4_-_28158335 | 0.37 |
ENSDART00000134605
|
gramd4a
|
GRAM domain containing 4a |
| chr2_-_13691834 | 0.37 |
ENSDART00000186570
|
CABZ01044235.1
|
|
| chr25_+_11281970 | 0.36 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr2_-_14587224 | 0.36 |
ENSDART00000085631
|
pde4bb
|
phosphodiesterase 4B, cAMP-specific b |
| chr1_+_34181581 | 0.36 |
ENSDART00000146042
|
epha6
|
eph receptor A6 |
| chr7_-_72337942 | 0.36 |
ENSDART00000162367
|
muc5.3
|
mucin 5.3 |
| chr13_+_2538636 | 0.35 |
ENSDART00000168927
|
plpp4
|
phospholipid phosphatase 4 |
| chr23_+_39963599 | 0.35 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr22_-_20379045 | 0.35 |
ENSDART00000183511
|
zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr17_+_39242437 | 0.35 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr2_-_24444838 | 0.35 |
ENSDART00000147885
ENSDART00000164720 |
kcnn1a
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1a |
| chr13_+_27232694 | 0.35 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr7_-_2047639 | 0.35 |
ENSDART00000173892
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr17_+_43486436 | 0.35 |
ENSDART00000023953
ENSDART00000149041 |
reep1
|
receptor accessory protein 1 |
| chr7_+_60551133 | 0.34 |
ENSDART00000148038
|
lrfn4b
|
leucine rich repeat and fibronectin type III domain containing 4b |
| chr9_+_17982737 | 0.34 |
ENSDART00000192569
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr13_+_23132666 | 0.34 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr8_-_25569920 | 0.34 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr17_+_12408405 | 0.34 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr15_-_45538773 | 0.34 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr8_-_15991801 | 0.34 |
ENSDART00000132005
|
agbl4
|
ATP/GTP binding protein-like 4 |
| chr8_+_9715010 | 0.34 |
ENSDART00000139414
|
gripap1
|
GRIP1 associated protein 1 |
| chr5_+_22633188 | 0.33 |
ENSDART00000128781
|
mtnr1c
|
melatonin receptor 1C |
| chr14_+_11909966 | 0.33 |
ENSDART00000171829
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr17_-_51262430 | 0.33 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr3_-_23406964 | 0.33 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr24_+_9178064 | 0.33 |
ENSDART00000142971
|
dlgap1b
|
discs, large (Drosophila) homolog-associated protein 1b |
| chr1_-_55196103 | 0.33 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr9_-_3034064 | 0.33 |
ENSDART00000115138
ENSDART00000144046 |
rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr22_-_14475927 | 0.33 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr17_-_13072334 | 0.33 |
ENSDART00000159598
|
CU469462.1
|
|
| chr23_-_31266586 | 0.32 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr7_-_32599669 | 0.32 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr14_-_2267515 | 0.32 |
ENSDART00000180988
ENSDART00000130712 |
pcdh2aa15
|
protocadherin 2 alpha a 15 |
| chr16_-_21591533 | 0.32 |
ENSDART00000159870
|
ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr6_+_40591149 | 0.32 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr18_+_36786842 | 0.32 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr14_-_14607855 | 0.32 |
ENSDART00000162322
|
rab9b
|
RAB9B, member RAS oncogene family |
| chr15_-_18432673 | 0.31 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr15_+_42397125 | 0.31 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr18_+_7283283 | 0.31 |
ENSDART00000141493
|
ANO2 (1 of many)
|
si:ch73-86n2.1 |
| chr14_+_6605675 | 0.31 |
ENSDART00000143179
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr3_-_42016693 | 0.31 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr14_-_2209742 | 0.31 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr21_-_2287589 | 0.31 |
ENSDART00000161554
|
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr3_+_6639796 | 0.30 |
ENSDART00000162557
|
cygb1
|
cytoglobin 1 |
| chr5_+_62374092 | 0.30 |
ENSDART00000082965
|
CU928117.1
|
|
| chr6_-_11136786 | 0.30 |
ENSDART00000186522
|
myo10l1
|
myosin X-like 1 |
| chr3_+_12748038 | 0.30 |
ENSDART00000181174
|
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr10_-_22249444 | 0.30 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr17_+_27176243 | 0.30 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr7_+_57725708 | 0.30 |
ENSDART00000056466
ENSDART00000142259 ENSDART00000166198 |
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr19_+_9186175 | 0.30 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr17_-_2386569 | 0.30 |
ENSDART00000121614
|
PLCB2
|
phospholipase C beta 2 |
| chr11_-_25853212 | 0.30 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr3_+_15773991 | 0.30 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr11_+_23760470 | 0.30 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr8_-_25327809 | 0.30 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr22_+_10129788 | 0.29 |
ENSDART00000045752
|
tbc1d20
|
TBC1 domain family, member 20 |
| chr2_+_5843694 | 0.29 |
ENSDART00000182364
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr14_-_33872092 | 0.29 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr4_-_8093753 | 0.29 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr15_+_30158652 | 0.29 |
ENSDART00000190682
|
nlk2
|
nemo-like kinase, type 2 |
| chr4_+_20812900 | 0.29 |
ENSDART00000005847
|
nav3
|
neuron navigator 3 |
| chr14_-_2217285 | 0.29 |
ENSDART00000157949
ENSDART00000166150 ENSDART00000054891 ENSDART00000183268 |
pcdh2ab2
pcdh2ab2
|
protocadherin 2 alpha b2 protocadherin 2 alpha b2 |
| chr1_+_19602389 | 0.29 |
ENSDART00000088933
ENSDART00000141579 ENSDART00000111555 |
fbxo10
|
F-box protein 10 |
| chr5_-_33215261 | 0.29 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr19_-_6083761 | 0.28 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr16_-_12173399 | 0.28 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr11_+_29770966 | 0.28 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr23_+_2760573 | 0.28 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr1_-_43897831 | 0.28 |
ENSDART00000048225
|
si:dkey-22i16.2
|
si:dkey-22i16.2 |
| chr20_-_37629084 | 0.28 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr15_+_19915345 | 0.28 |
ENSDART00000114267
|
map6b
|
microtubule-associated protein 6b |
| chr9_-_16109001 | 0.28 |
ENSDART00000053473
|
upp2
|
uridine phosphorylase 2 |
| chr7_+_38962207 | 0.28 |
ENSDART00000173565
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr11_-_40742424 | 0.27 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr2_+_55914699 | 0.27 |
ENSDART00000157905
|
atcayb
|
ataxia, cerebellar, Cayman type b |
| chr15_-_40267485 | 0.27 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr10_-_28519505 | 0.27 |
ENSDART00000137964
|
bbx
|
bobby sox homolog (Drosophila) |
| chr12_+_1286642 | 0.27 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr20_+_22067337 | 0.27 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr3_+_15776446 | 0.27 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr25_-_4530085 | 0.26 |
ENSDART00000144907
|
pidd1
|
p53-induced death domain protein 1 |
| chr2_+_11028923 | 0.26 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr12_-_11560794 | 0.26 |
ENSDART00000149098
ENSDART00000169975 |
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr21_+_37357578 | 0.26 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr9_-_53537989 | 0.26 |
ENSDART00000114022
|
slitrk5b
|
SLIT and NTRK-like family, member 5b |
| chr24_-_21512425 | 0.26 |
ENSDART00000184857
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr7_+_57109214 | 0.26 |
ENSDART00000135068
ENSDART00000098412 |
enosf1
|
enolase superfamily member 1 |
| chr19_+_22727940 | 0.25 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr24_-_1151334 | 0.25 |
ENSDART00000039700
ENSDART00000177356 |
itgb1a
|
integrin, beta 1a |
| chr25_+_10762016 | 0.25 |
ENSDART00000131231
|
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr8_-_19649617 | 0.25 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr1_-_54191626 | 0.25 |
ENSDART00000062941
|
nthl1
|
nth-like DNA glycosylase 1 |
| chr8_-_24252933 | 0.25 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx5a+tbx5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.2 | 0.7 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 0.8 | GO:2000392 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.2 | 0.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.6 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.1 | 0.3 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 0.3 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.4 | GO:0048241 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.5 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.4 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.1 | 0.4 | GO:0034164 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.5 | GO:0051969 | regulation of transmission of nerve impulse(GO:0051969) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.2 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 0.2 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.2 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.8 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 0.5 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.8 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.4 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.2 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 0.8 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.4 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.3 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.2 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.1 | 0.2 | GO:0002420 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 0.2 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 1.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.4 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.6 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.5 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.2 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.0 | 0.2 | GO:0061032 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.0 | 0.4 | GO:0043490 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.2 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.5 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.4 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.3 | GO:0010138 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:0071423 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) C4-dicarboxylate transport(GO:0015740) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.3 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 1.2 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.0 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.1 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) bone remodeling(GO:0046849) |
| 0.0 | 0.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.3 | GO:1903288 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.0 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.3 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.3 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.6 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.0 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.2 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.7 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.3 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 0.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.4 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 0.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 1.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.3 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 0.7 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.7 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 1.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.1 | 0.8 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.2 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.6 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.5 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.2 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.1 | 0.3 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.2 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 0.3 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.0 | 0.8 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.3 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.5 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.2 | GO:0008515 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.2 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0005343 | organic acid:sodium symporter activity(GO:0005343) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.5 | GO:0017022 | myosin binding(GO:0017022) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.7 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.1 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 1.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |