Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
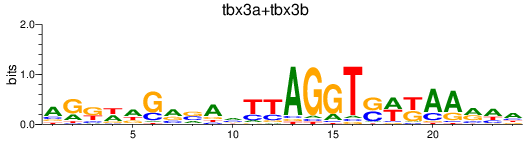
Results for tbx3a+tbx3b
Z-value: 0.94
Transcription factors associated with tbx3a+tbx3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx3a
|
ENSDARG00000002216 | T-box transcription factor 3a |
|
tbx3b
|
ENSDARG00000061509 | T-box transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx3b | dr11_v1_chr5_+_26913120_26913120 | 0.86 | 2.1e-06 | Click! |
| tbx3a | dr11_v1_chr5_-_72324371_72324371 | 0.63 | 4.1e-03 | Click! |
Activity profile of tbx3a+tbx3b motif
Sorted Z-values of tbx3a+tbx3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_26532167 | 3.53 |
ENSDART00000061914
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like |
| chr12_+_27141140 | 3.40 |
ENSDART00000136415
|
hoxb1b
|
homeobox B1b |
| chr11_-_23219367 | 3.03 |
ENSDART00000003646
|
optc
|
opticin |
| chr18_+_37272568 | 2.46 |
ENSDART00000132749
|
tmem123
|
transmembrane protein 123 |
| chr3_+_37790351 | 2.41 |
ENSDART00000151506
|
si:dkey-260c8.8
|
si:dkey-260c8.8 |
| chr19_+_7424347 | 2.33 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr4_+_15006217 | 2.00 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr4_+_14981854 | 1.80 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr1_+_25650917 | 1.72 |
ENSDART00000054235
|
plrg1
|
pleiotropic regulator 1 |
| chr21_+_21611867 | 1.53 |
ENSDART00000189148
|
b9d2
|
B9 domain containing 2 |
| chr5_+_29803380 | 1.50 |
ENSDART00000005263
ENSDART00000137558 ENSDART00000146963 ENSDART00000134900 |
usf1l
|
upstream transcription factor 1, like |
| chr13_-_11984867 | 1.49 |
ENSDART00000157538
|
npm3
|
nucleophosmin/nucleoplasmin, 3 |
| chr19_-_18127629 | 1.46 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr23_-_20345473 | 1.45 |
ENSDART00000140935
|
si:rp71-17i16.6
|
si:rp71-17i16.6 |
| chr18_-_40913294 | 1.43 |
ENSDART00000059196
ENSDART00000098878 |
polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I |
| chr14_+_3516567 | 1.43 |
ENSDART00000171153
ENSDART00000158416 |
fnta
|
farnesyltransferase, CAAX box, alpha |
| chr5_+_69278089 | 1.43 |
ENSDART00000136656
|
selenom
|
selenoprotein M |
| chr15_-_41677689 | 1.40 |
ENSDART00000187063
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr1_+_218524 | 1.39 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr17_+_24604373 | 1.33 |
ENSDART00000082193
|
zmpste24
|
zinc metallopeptidase, STE24 homolog |
| chr7_+_29163762 | 1.33 |
ENSDART00000173762
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr19_-_18127808 | 1.33 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr4_-_5775507 | 1.31 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr8_-_46897734 | 1.31 |
ENSDART00000138125
|
hes2.2
|
hes family bHLH transcription factor 2, tandem duplicate 2 |
| chr23_-_19831739 | 1.31 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr2_-_37103622 | 1.29 |
ENSDART00000137849
|
zgc:101744
|
zgc:101744 |
| chr11_+_11271959 | 1.28 |
ENSDART00000190678
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr12_+_31537604 | 1.27 |
ENSDART00000153340
|
si:ch73-205h11.1
|
si:ch73-205h11.1 |
| chr23_+_2421689 | 1.26 |
ENSDART00000180200
|
tcp1
|
t-complex 1 |
| chr17_-_27273296 | 1.22 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr21_-_27195256 | 1.18 |
ENSDART00000133152
ENSDART00000065401 |
zgc:110782
|
zgc:110782 |
| chr4_+_15899668 | 1.17 |
ENSDART00000101592
|
col7a1l
|
collagen type VII alpha 1-like |
| chr13_+_33304187 | 1.15 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr2_+_23482798 | 1.15 |
ENSDART00000187448
|
BX470197.1
|
|
| chr12_-_26415499 | 1.15 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr14_+_20911310 | 1.13 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr20_-_34164278 | 1.09 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr16_-_47301376 | 1.09 |
ENSDART00000169697
|
mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr13_-_51903150 | 1.02 |
ENSDART00000090644
|
mrpl2
|
mitochondrial ribosomal protein L2 |
| chr6_-_55399214 | 1.01 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr8_+_15251448 | 1.00 |
ENSDART00000063717
|
zgc:171480
|
zgc:171480 |
| chr2_+_42005475 | 0.97 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr16_-_14587332 | 0.97 |
ENSDART00000012479
|
dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chr21_+_21612214 | 0.94 |
ENSDART00000008099
|
b9d2
|
B9 domain containing 2 |
| chr13_+_25486608 | 0.93 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr22_-_9157364 | 0.91 |
ENSDART00000182762
|
si:ch211-213a13.5
|
si:ch211-213a13.5 |
| chr18_-_21748966 | 0.91 |
ENSDART00000181910
|
pskh1
|
protein serine kinase H1 |
| chr5_-_17601759 | 0.89 |
ENSDART00000138387
|
si:ch211-130h14.6
|
si:ch211-130h14.6 |
| chr2_-_21438492 | 0.88 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr6_-_40651944 | 0.86 |
ENSDART00000187423
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr11_-_25461336 | 0.84 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr3_-_36690348 | 0.83 |
ENSDART00000192513
|
myh11b
|
myosin, heavy chain 11b, smooth muscle |
| chr17_-_50071748 | 0.82 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr8_-_50979047 | 0.80 |
ENSDART00000184788
ENSDART00000180906 |
zgc:91909
|
zgc:91909 |
| chr7_+_17106160 | 0.78 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr8_+_7854130 | 0.77 |
ENSDART00000165575
|
cxxc1a
|
CXXC finger protein 1a |
| chr16_+_23331633 | 0.76 |
ENSDART00000187455
|
krtcap2
|
keratinocyte associated protein 2 |
| chr19_-_23674904 | 0.75 |
ENSDART00000187300
|
BX927234.2
|
|
| chr4_-_60518031 | 0.74 |
ENSDART00000157489
|
si:dkey-211i20.4
|
si:dkey-211i20.4 |
| chr5_+_29803702 | 0.74 |
ENSDART00000147569
|
usf1l
|
upstream transcription factor 1, like |
| chr25_-_27735290 | 0.73 |
ENSDART00000140663
|
zgc:153935
|
zgc:153935 |
| chr9_+_30108641 | 0.73 |
ENSDART00000060174
|
jagn1a
|
jagunal homolog 1a |
| chr14_-_28052474 | 0.71 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr3_+_28831450 | 0.69 |
ENSDART00000055422
|
flr
|
fleer |
| chr10_+_26652859 | 0.68 |
ENSDART00000079174
|
htatsf1
|
HIV-1 Tat specific factor 1 |
| chr16_+_32184485 | 0.67 |
ENSDART00000084009
|
zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chr2_+_43895103 | 0.66 |
ENSDART00000007074
|
gbp3
|
guanylate binding protein 3 |
| chr1_+_54901028 | 0.66 |
ENSDART00000137352
|
zfyve27
|
zinc finger, FYVE domain containing 27 |
| chr5_-_10236599 | 0.64 |
ENSDART00000099834
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr16_-_41990421 | 0.64 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr21_+_22423286 | 0.63 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr19_-_10810006 | 0.62 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr21_+_7332963 | 0.61 |
ENSDART00000169834
|
AP3B1
|
adaptor related protein complex 3 beta 1 subunit |
| chr7_+_56472585 | 0.60 |
ENSDART00000135259
ENSDART00000073596 |
ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr22_-_5441893 | 0.60 |
ENSDART00000161421
|
zgc:194627
|
zgc:194627 |
| chr4_+_77908076 | 0.53 |
ENSDART00000168811
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr21_-_33478164 | 0.53 |
ENSDART00000191542
|
si:ch73-42p12.2
|
si:ch73-42p12.2 |
| chr12_+_18663154 | 0.52 |
ENSDART00000057918
|
si:ch211-147h1.4
|
si:ch211-147h1.4 |
| chr4_+_77681389 | 0.52 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr13_+_30163515 | 0.52 |
ENSDART00000040926
|
eif4ebp2
|
eukaryotic translation initiation factor 4E binding protein 2 |
| chr2_+_23677179 | 0.49 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr5_-_51148298 | 0.49 |
ENSDART00000042420
|
ccdc180
|
coiled-coil domain containing 180 |
| chr14_+_23687678 | 0.48 |
ENSDART00000002469
|
hspa4b
|
heat shock protein 4b |
| chr3_-_33967767 | 0.47 |
ENSDART00000151493
ENSDART00000151160 |
ighv1-4
|
immunoglobulin heavy variable 1-4 |
| chr18_-_7097403 | 0.47 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr24_-_23942722 | 0.46 |
ENSDART00000080810
|
arxa
|
aristaless related homeobox a |
| chr17_+_8175998 | 0.43 |
ENSDART00000131200
|
myct1b
|
myc target 1b |
| chr10_-_15048781 | 0.42 |
ENSDART00000038401
ENSDART00000155674 |
si:ch211-95j8.2
|
si:ch211-95j8.2 |
| chr12_+_20583552 | 0.41 |
ENSDART00000170035
|
arsg
|
arylsulfatase G |
| chr16_-_32184460 | 0.40 |
ENSDART00000102027
|
kpna5
|
karyopherin alpha 5 (importin alpha 6) |
| chr22_+_344763 | 0.40 |
ENSDART00000181934
|
CU914780.1
|
|
| chr17_-_27200634 | 0.39 |
ENSDART00000185332
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr22_+_9294336 | 0.39 |
ENSDART00000193694
|
si:ch211-250k18.7
|
si:ch211-250k18.7 |
| chr17_-_53329704 | 0.38 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr22_+_2451732 | 0.37 |
ENSDART00000164419
|
znf1177
|
zinc finger protein 1177 |
| chr7_-_33960170 | 0.37 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr25_-_27819838 | 0.36 |
ENSDART00000067106
|
lmod2a
|
leiomodin 2 (cardiac) a |
| chr6_-_1514767 | 0.35 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr1_-_2453174 | 0.35 |
ENSDART00000055779
ENSDART00000152555 |
ggact.2
ggact.3
|
gamma-glutamylamine cyclotransferase, tandem duplicate 2 gamma-glutamylamine cyclotransferase, tandem duplicate 3 |
| chr23_-_5683147 | 0.35 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr5_-_60159116 | 0.34 |
ENSDART00000147675
|
si:dkey-280e8.1
|
si:dkey-280e8.1 |
| chr15_-_5178899 | 0.33 |
ENSDART00000132148
|
or126-4
|
odorant receptor, family E, subfamily 126, member 4 |
| chr2_-_38992304 | 0.33 |
ENSDART00000114085
ENSDART00000146812 |
si:ch211-119o8.6
|
si:ch211-119o8.6 |
| chr9_-_105135 | 0.33 |
ENSDART00000180126
|
FQ377903.3
|
|
| chr22_+_567733 | 0.32 |
ENSDART00000171036
|
usp49
|
ubiquitin specific peptidase 49 |
| chr25_-_15504559 | 0.32 |
ENSDART00000139294
|
BX323543.5
|
|
| chr12_+_36109071 | 0.32 |
ENSDART00000171268
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr7_-_3836635 | 0.30 |
ENSDART00000104350
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr5_+_69856153 | 0.29 |
ENSDART00000124128
ENSDART00000073663 |
ugt2a6
|
UDP glucuronosyltransferase 2 family, polypeptide A6 |
| chr4_-_16001118 | 0.27 |
ENSDART00000041070
ENSDART00000125389 |
mest
|
mesoderm specific transcript |
| chr19_+_18493782 | 0.27 |
ENSDART00000160992
|
fkbp10a
|
FK506 binding protein 10a |
| chr21_+_25688388 | 0.26 |
ENSDART00000125709
|
bicdl2
|
bicaudal-D-related protein 2 |
| chr9_-_29321625 | 0.25 |
ENSDART00000158689
ENSDART00000014047 ENSDART00000122602 |
pth2r
|
parathyroid hormone 2 receptor |
| chr25_-_15512819 | 0.25 |
ENSDART00000142684
|
si:dkeyp-67e1.2
|
si:dkeyp-67e1.2 |
| chr7_-_3826794 | 0.25 |
ENSDART00000064229
ENSDART00000142138 |
si:dkey-88n24.10
|
si:dkey-88n24.10 |
| chr19_+_9212031 | 0.24 |
ENSDART00000052930
|
ndufv1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1 |
| chr9_+_50001746 | 0.24 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr18_+_32916351 | 0.23 |
ENSDART00000170387
ENSDART00000161553 |
olfcg2
|
olfactory receptor C family, g2 |
| chr14_-_8903435 | 0.23 |
ENSDART00000160584
|
zgc:153681
|
zgc:153681 |
| chr8_-_32894203 | 0.22 |
ENSDART00000147822
|
si:dkey-56i24.1
|
si:dkey-56i24.1 |
| chr7_+_65121743 | 0.22 |
ENSDART00000108471
|
ipo7
|
importin 7 |
| chr16_-_32672883 | 0.20 |
ENSDART00000124515
ENSDART00000190920 ENSDART00000188776 |
pnisr
|
PNN-interacting serine/arginine-rich protein |
| chr2_-_51611344 | 0.20 |
ENSDART00000158137
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr7_+_2236317 | 0.19 |
ENSDART00000075859
|
zgc:172065
|
zgc:172065 |
| chr5_-_38197080 | 0.18 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr6_+_9793791 | 0.18 |
ENSDART00000149896
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr24_-_26479841 | 0.18 |
ENSDART00000079984
|
rpl22l1
|
ribosomal protein L22-like 1 |
| chr4_+_51542447 | 0.17 |
ENSDART00000163814
|
si:dkey-165e24.1
|
si:dkey-165e24.1 |
| chr12_+_20506197 | 0.17 |
ENSDART00000153010
|
si:zfos-754c12.2
|
si:zfos-754c12.2 |
| chr8_+_26818446 | 0.17 |
ENSDART00000134987
ENSDART00000138835 |
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr8_-_25336589 | 0.17 |
ENSDART00000009682
|
atp5pb
|
ATP synthase peripheral stalk-membrane subunit b |
| chr4_+_62598975 | 0.17 |
ENSDART00000163548
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr20_-_874807 | 0.16 |
ENSDART00000020506
|
snx14
|
sorting nexin 14 |
| chr7_-_3836896 | 0.16 |
ENSDART00000136227
|
si:dkey-88n24.8
|
si:dkey-88n24.8 |
| chr24_-_25184553 | 0.14 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr8_+_20918207 | 0.14 |
ENSDART00000144039
|
si:ch73-196i15.5
|
si:ch73-196i15.5 |
| chr12_-_26491464 | 0.14 |
ENSDART00000153361
|
si:dkey-287g12.6
|
si:dkey-287g12.6 |
| chr13_+_33462232 | 0.14 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr23_-_6722101 | 0.14 |
ENSDART00000157828
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr16_-_50918908 | 0.14 |
ENSDART00000108744
|
CU855821.1
|
|
| chr3_+_5297493 | 0.13 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
| chr23_-_15090782 | 0.12 |
ENSDART00000133624
|
si:ch211-218g4.2
|
si:ch211-218g4.2 |
| chr18_+_29145681 | 0.12 |
ENSDART00000089031
ENSDART00000193336 |
ppfibp2a
|
PTPRF interacting protein, binding protein 2a (liprin beta 2) |
| chr2_+_27403300 | 0.12 |
ENSDART00000099180
|
elovl8a
|
ELOVL fatty acid elongase 8a |
| chr7_+_13756374 | 0.10 |
ENSDART00000180808
|
rasl12
|
RAS-like, family 12 |
| chr5_-_20491198 | 0.10 |
ENSDART00000183051
ENSDART00000144232 |
ficd
|
FIC domain containing |
| chr21_+_21655755 | 0.10 |
ENSDART00000079524
|
or125-8
|
odorant receptor, family E, subfamily 125, member 8 |
| chr7_+_24889783 | 0.09 |
ENSDART00000005329
ENSDART00000159955 |
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr4_-_36792144 | 0.07 |
ENSDART00000159147
|
si:dkeyp-87d1.1
|
si:dkeyp-87d1.1 |
| chr18_+_17538165 | 0.06 |
ENSDART00000147839
|
nup93
|
nucleoporin 93 |
| chr6_+_15250672 | 0.05 |
ENSDART00000155951
|
si:ch73-23l24.1
|
si:ch73-23l24.1 |
| chr5_-_67783593 | 0.04 |
ENSDART00000010934
|
casr
|
calcium-sensing receptor |
| chr4_+_76431531 | 0.03 |
ENSDART00000174278
|
FP074874.1
|
|
| chr15_+_23523337 | 0.03 |
ENSDART00000182306
ENSDART00000185545 ENSDART00000152517 |
si:dkey-182i3.9
|
si:dkey-182i3.9 |
| chr6_+_9793495 | 0.02 |
ENSDART00000108524
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr3_-_48612078 | 0.01 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr4_+_332709 | 0.01 |
ENSDART00000067486
|
tulp4a
|
tubby like protein 4a |
| chr9_+_34334156 | 0.01 |
ENSDART00000144272
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr11_+_44226200 | 0.01 |
ENSDART00000191417
|
gnb4b
|
guanine nucleotide binding protein (G protein), beta polypeptide 4b |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx3a+tbx3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.5 | 3.4 | GO:0021570 | rhombomere 4 development(GO:0021570) rhombomere 4 morphogenesis(GO:0021661) |
| 0.4 | 1.3 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.4 | 1.7 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.4 | 3.2 | GO:0036372 | opsin transport(GO:0036372) |
| 0.4 | 1.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.4 | 1.4 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 1.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.3 | 0.9 | GO:0010662 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 0.8 | GO:0042543 | protein N-linked glycosylation via arginine(GO:0042543) |
| 0.2 | 0.7 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.2 | 3.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 1.3 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 2.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.7 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.3 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 1.2 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.1 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 1.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.8 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.0 | 0.8 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.1 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 1.3 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.6 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.0 | 0.5 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 1.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.3 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.9 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.3 | 1.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.2 | 1.7 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 0.7 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 0.6 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 2.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.7 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 1.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.4 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 2.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 1.3 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 2.5 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.5 | 1.4 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.5 | 1.8 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.3 | 1.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.2 | 0.7 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.2 | 0.7 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 1.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 2.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.9 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 1.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.4 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.8 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.6 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.6 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:0010181 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) FMN binding(GO:0010181) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.1 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.4 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 2.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.1 | 1.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |