Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
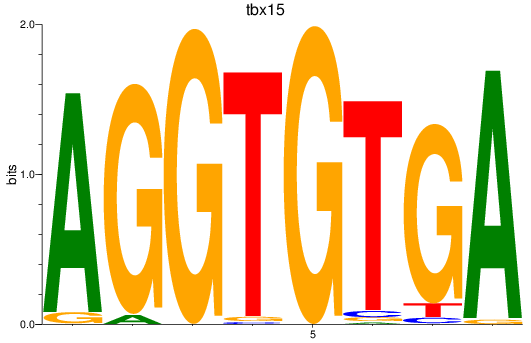
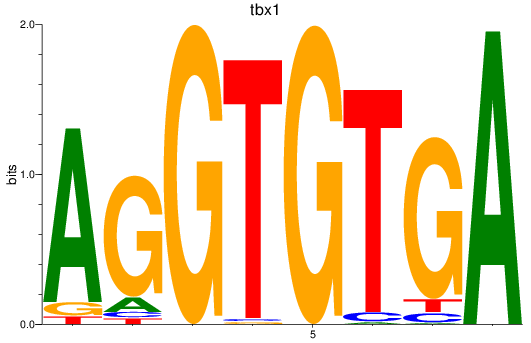
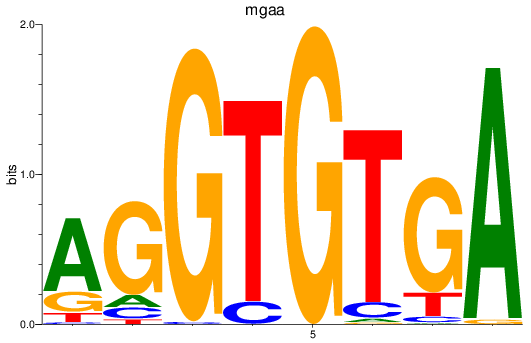
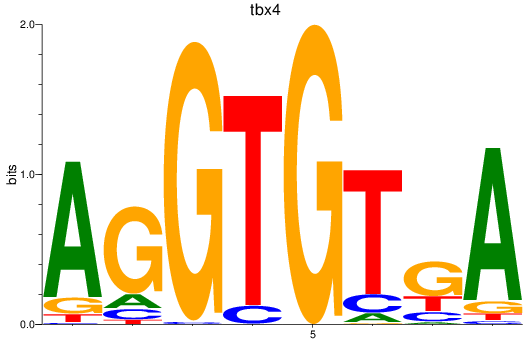
Results for tbx15_tbx1_mgaa_tbx4
Z-value: 1.78
Transcription factors associated with tbx15_tbx1_mgaa_tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx15
|
ENSDARG00000002582 | T-box transcription factor 15 |
|
tbx1
|
ENSDARG00000031891 | T-box transcription factor 1 |
|
mgaa
|
ENSDARG00000078784 | MAX dimerization protein MGA a |
|
tbx4
|
ENSDARG00000030058 | T-box transcription factor 4 |
|
tbx4
|
ENSDARG00000113067 | T-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mgaa | dr11_v1_chr17_+_10566490_10566495 | 0.88 | 6.7e-07 | Click! |
| tbx1 | dr11_v1_chr5_+_15203421_15203421 | -0.60 | 6.2e-03 | Click! |
| tbx15 | dr11_v1_chr9_-_21067971_21067971 | -0.55 | 1.6e-02 | Click! |
| tbx4 | dr11_v1_chr15_+_27387555_27387555 | 0.28 | 2.4e-01 | Click! |
Activity profile of tbx15_tbx1_mgaa_tbx4 motif
Sorted Z-values of tbx15_tbx1_mgaa_tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_38638809 | 6.63 |
ENSDART00000144341
|
c6ast4
|
six-cysteine containing astacin protease 4 |
| chr3_-_53465223 | 5.62 |
ENSDART00000057123
ENSDART00000125515 ENSDART00000143096 |
nr5a5
|
nuclear receptor subfamily 5, group A, member 5 |
| chr9_-_42873700 | 4.65 |
ENSDART00000125953
|
ttn.1
|
titin, tandem duplicate 1 |
| chr21_-_43550120 | 4.51 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr16_-_13613475 | 4.26 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr9_-_43073960 | 4.22 |
ENSDART00000059460
|
ttn.2
|
titin, tandem duplicate 2 |
| chr21_-_3700334 | 3.91 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr13_+_44495063 | 3.58 |
ENSDART00000169591
|
CU694264.1
|
|
| chr22_-_14475927 | 3.16 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr19_-_35229336 | 3.15 |
ENSDART00000054274
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr13_-_24906307 | 3.09 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr5_+_22791686 | 2.99 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr21_+_44300689 | 2.94 |
ENSDART00000186298
ENSDART00000142810 |
gabra3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3 |
| chr18_-_51015718 | 2.92 |
ENSDART00000190698
|
LO018598.1
|
|
| chr11_+_1628538 | 2.91 |
ENSDART00000154967
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr12_-_31457301 | 2.90 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr13_-_36264861 | 2.85 |
ENSDART00000100204
|
slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr5_+_37837245 | 2.79 |
ENSDART00000171617
|
epd
|
ependymin |
| chr23_+_28582865 | 2.74 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr4_+_11384891 | 2.71 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr22_+_26600834 | 2.66 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr11_+_25583950 | 2.62 |
ENSDART00000111961
|
ccdc120
|
coiled-coil domain containing 120 |
| chr6_+_40587551 | 2.60 |
ENSDART00000155554
|
frs3
|
fibroblast growth factor receptor substrate 3 |
| chr14_+_45406299 | 2.59 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr13_+_38817871 | 2.58 |
ENSDART00000187708
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr21_-_10488379 | 2.57 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr22_-_12160283 | 2.57 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr23_-_46020226 | 2.47 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr9_-_32912638 | 2.39 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr2_-_5776030 | 2.37 |
ENSDART00000133851
|
nyap2b
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2b |
| chr8_+_10862353 | 2.35 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr17_+_27176243 | 2.32 |
ENSDART00000162527
|
si:ch211-160f23.7
|
si:ch211-160f23.7 |
| chr16_-_13109749 | 2.31 |
ENSDART00000142610
|
prkcg
|
protein kinase C, gamma |
| chr23_+_40604951 | 2.25 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr15_+_42397125 | 2.25 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr3_-_33901483 | 2.21 |
ENSDART00000144774
ENSDART00000138765 |
cacna1aa
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, a |
| chr17_-_3986236 | 2.21 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr5_+_19343880 | 2.20 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr11_-_37880492 | 2.16 |
ENSDART00000102868
|
etnk2
|
ethanolamine kinase 2 |
| chr14_+_6605675 | 2.15 |
ENSDART00000143179
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr24_-_36095526 | 2.13 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr7_+_11197022 | 2.12 |
ENSDART00000185236
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr16_-_12173399 | 2.08 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr17_-_15498275 | 2.04 |
ENSDART00000156905
ENSDART00000080661 |
si:ch211-266g18.10
|
si:ch211-266g18.10 |
| chr9_-_21970067 | 2.03 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr17_-_20394566 | 2.03 |
ENSDART00000154667
|
sorcs3b
|
sortilin related VPS10 domain containing receptor 3b |
| chr15_-_10455438 | 2.02 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr3_+_23488652 | 1.97 |
ENSDART00000126282
|
nr1d1
|
nuclear receptor subfamily 1, group d, member 1 |
| chr24_+_35975398 | 1.96 |
ENSDART00000173058
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr1_-_12126535 | 1.94 |
ENSDART00000164817
ENSDART00000015251 |
mttp
|
microsomal triglyceride transfer protein |
| chr8_+_8459192 | 1.92 |
ENSDART00000140942
ENSDART00000014939 |
comta
|
catechol-O-methyltransferase a |
| chr23_+_37482727 | 1.89 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr7_-_38612230 | 1.89 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr11_+_23933016 | 1.88 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr1_-_40341306 | 1.87 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr2_-_32501501 | 1.87 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr7_+_6990510 | 1.87 |
ENSDART00000138948
|
ccs
|
copper chaperone for superoxide dismutase |
| chr3_-_39171968 | 1.85 |
ENSDART00000154494
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr15_-_43164591 | 1.85 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr16_+_8716800 | 1.85 |
ENSDART00000124693
ENSDART00000181961 |
cabz01093075.1
|
cabz01093075.1 |
| chr19_-_22621991 | 1.84 |
ENSDART00000175106
|
pleca
|
plectin a |
| chr25_-_30429607 | 1.82 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr7_-_51476276 | 1.81 |
ENSDART00000082464
|
nhsl2
|
NHS-like 2 |
| chr1_-_714626 | 1.78 |
ENSDART00000161072
|
adamts5
|
ADAM metallopeptidase with thrombospondin type 1 motif 5 |
| chr22_+_19290199 | 1.77 |
ENSDART00000148173
|
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr6_+_56141852 | 1.76 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr23_-_31266586 | 1.75 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr19_+_32456974 | 1.75 |
ENSDART00000088265
|
atxn1a
|
ataxin 1a |
| chr24_-_32150276 | 1.73 |
ENSDART00000166212
|
cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr17_+_3124129 | 1.71 |
ENSDART00000155323
|
zgc:136872
|
zgc:136872 |
| chr21_-_43949208 | 1.70 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr8_-_4508248 | 1.70 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr13_-_27653679 | 1.69 |
ENSDART00000142568
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr14_+_2487672 | 1.69 |
ENSDART00000170629
ENSDART00000123063 |
fgf18a
|
fibroblast growth factor 18a |
| chr8_-_19246342 | 1.69 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr12_-_26383242 | 1.68 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr14_+_1365097 | 1.68 |
ENSDART00000083797
ENSDART00000176087 |
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr13_-_51846224 | 1.66 |
ENSDART00000184663
|
LT631684.2
|
|
| chr4_-_63848305 | 1.66 |
ENSDART00000097308
|
BX901974.1
|
|
| chr19_+_14921000 | 1.64 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr17_+_39242437 | 1.63 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr13_-_21739142 | 1.62 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr10_+_32066355 | 1.60 |
ENSDART00000062311
|
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr5_+_26075230 | 1.59 |
ENSDART00000098473
|
klf9
|
Kruppel-like factor 9 |
| chr5_+_22633188 | 1.59 |
ENSDART00000128781
|
mtnr1c
|
melatonin receptor 1C |
| chr7_+_32369463 | 1.59 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr2_-_53500424 | 1.58 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr14_-_31059218 | 1.57 |
ENSDART00000111691
ENSDART00000021379 ENSDART00000113479 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr22_-_20575679 | 1.56 |
ENSDART00000089033
|
lingo3a
|
leucine rich repeat and Ig domain containing 3a |
| chr5_-_33215261 | 1.55 |
ENSDART00000097935
ENSDART00000134777 |
si:dkey-226m8.10
|
si:dkey-226m8.10 |
| chr10_+_32066537 | 1.55 |
ENSDART00000124166
|
si:dkey-250d21.1
|
si:dkey-250d21.1 |
| chr5_-_37875636 | 1.55 |
ENSDART00000184674
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr7_+_65240227 | 1.54 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr21_-_2287589 | 1.54 |
ENSDART00000161554
|
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr5_-_4204580 | 1.53 |
ENSDART00000049197
ENSDART00000132130 |
si:ch211-283g2.1
|
si:ch211-283g2.1 |
| chr20_-_23946296 | 1.53 |
ENSDART00000143005
|
mdn1
|
midasin AAA ATPase 1 |
| chr12_-_5448993 | 1.52 |
ENSDART00000181802
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr11_+_41540862 | 1.49 |
ENSDART00000173210
|
kcnab2a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 a |
| chr15_+_40665310 | 1.49 |
ENSDART00000154187
ENSDART00000042082 |
fat3a
|
FAT atypical cadherin 3a |
| chr5_-_26118855 | 1.47 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr1_-_23110740 | 1.46 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr15_+_42599501 | 1.46 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr15_-_18432673 | 1.46 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr19_+_791538 | 1.44 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr11_+_29770966 | 1.44 |
ENSDART00000088624
ENSDART00000124471 |
rpgrb
|
retinitis pigmentosa GTPase regulator b |
| chr8_-_1838315 | 1.43 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr20_-_7128612 | 1.43 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr23_+_27912079 | 1.42 |
ENSDART00000171859
|
BX539336.1
|
|
| chr25_+_37397031 | 1.42 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr18_-_8398972 | 1.41 |
ENSDART00000136512
|
BEND7
|
si:ch211-220f12.4 |
| chr19_+_7735157 | 1.41 |
ENSDART00000186717
|
tuft1b
|
tuftelin 1b |
| chr4_-_60049792 | 1.41 |
ENSDART00000158199
|
znf1033
|
zinc finger protein 1033 |
| chr7_-_32599669 | 1.39 |
ENSDART00000173752
|
kcna4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr7_+_7048245 | 1.38 |
ENSDART00000001649
|
actn3b
|
actinin alpha 3b |
| chr22_-_16275236 | 1.38 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr23_-_4855122 | 1.38 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr17_+_28340138 | 1.36 |
ENSDART00000033943
|
mdga2a
|
MAM domain containing glycosylphosphatidylinositol anchor 2a |
| chr17_-_26537928 | 1.35 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr22_+_19289970 | 1.35 |
ENSDART00000137976
ENSDART00000132386 |
si:dkey-21e2.15
|
si:dkey-21e2.15 |
| chr20_-_26846028 | 1.34 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr23_+_44263986 | 1.34 |
ENSDART00000149194
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr14_+_45925810 | 1.32 |
ENSDART00000189543
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr9_+_36356740 | 1.31 |
ENSDART00000139033
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr19_+_1005933 | 1.30 |
ENSDART00000191953
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr7_+_14005111 | 1.30 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr7_+_30867008 | 1.29 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr10_-_44411032 | 1.28 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr6_-_18199062 | 1.27 |
ENSDART00000167513
|
ppp1r27b
|
protein phosphatase 1, regulatory subunit 27b |
| chr10_-_20588787 | 1.26 |
ENSDART00000138045
ENSDART00000181885 ENSDART00000091115 |
nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr12_+_15290800 | 1.25 |
ENSDART00000145656
|
med1
|
mediator complex subunit 1 |
| chr7_+_6969909 | 1.25 |
ENSDART00000189886
|
actn3b
|
actinin alpha 3b |
| chr25_-_12788370 | 1.23 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr4_-_4795205 | 1.22 |
ENSDART00000039313
|
zgc:162331
|
zgc:162331 |
| chr13_-_24880525 | 1.21 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr8_-_25327809 | 1.21 |
ENSDART00000137242
|
eps8l3b
|
EPS8-like 3b |
| chr15_+_28482862 | 1.18 |
ENSDART00000015286
ENSDART00000154320 |
ankrd13b
|
ankyrin repeat domain 13B |
| chr25_+_32525131 | 1.18 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr12_+_32292564 | 1.18 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr5_+_60919378 | 1.17 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr13_+_40437550 | 1.16 |
ENSDART00000057090
ENSDART00000167859 |
got1
|
glutamic-oxaloacetic transaminase 1, soluble |
| chr16_-_29437373 | 1.16 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr6_+_58406014 | 1.16 |
ENSDART00000044241
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr23_+_21380079 | 1.15 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr2_+_15209676 | 1.14 |
ENSDART00000090760
|
abca4b
|
ATP-binding cassette, sub-family A (ABC1), member 4b |
| chr9_+_42095220 | 1.14 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr2_+_33368414 | 1.12 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr22_-_9667136 | 1.12 |
ENSDART00000181154
|
si:dkey-286j17.4
|
si:dkey-286j17.4 |
| chr20_+_41549735 | 1.11 |
ENSDART00000184235
|
fam184a
|
family with sequence similarity 184, member A |
| chr7_-_72295181 | 1.10 |
ENSDART00000180598
|
muc5.3
|
mucin 5.3 |
| chr20_-_52902693 | 1.10 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr3_-_30888415 | 1.08 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr20_-_51946052 | 1.08 |
ENSDART00000074325
|
dusp10
|
dual specificity phosphatase 10 |
| chr23_-_29003864 | 1.07 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr7_-_39378903 | 1.07 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr2_+_24203229 | 1.06 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr24_-_6158933 | 1.05 |
ENSDART00000021609
|
gad2
|
glutamate decarboxylase 2 |
| chr1_-_53625142 | 1.04 |
ENSDART00000166852
|
USP34
|
ubiquitin specific peptidase 34 |
| chr11_-_26832685 | 1.03 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr12_+_34770531 | 1.03 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr9_-_41323746 | 1.03 |
ENSDART00000140564
|
glsb
|
glutaminase b |
| chr4_+_77735212 | 1.03 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr8_+_10869183 | 1.03 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr17_-_27976031 | 1.01 |
ENSDART00000154829
|
ptafr
|
platelet-activating factor receptor |
| chr23_-_5032587 | 1.00 |
ENSDART00000163903
|
kcna2b
|
potassium voltage-gated channel, shaker-related subfamily, member 2b |
| chr13_-_31888487 | 1.00 |
ENSDART00000145938
|
syt14a
|
synaptotagmin XIVa |
| chr8_-_44926077 | 1.00 |
ENSDART00000084417
|
timm17b
|
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
| chr22_-_3152357 | 0.99 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr14_-_26177156 | 0.98 |
ENSDART00000014149
|
fat2
|
FAT atypical cadherin 2 |
| chr16_-_35532937 | 0.98 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr9_-_14992730 | 0.98 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr8_-_6918290 | 0.98 |
ENSDART00000138259
ENSDART00000142496 |
asb6
|
ankyrin repeat and SOCS box containing 6 |
| chr7_+_31120766 | 0.98 |
ENSDART00000173703
|
tjp1a
|
tight junction protein 1a |
| chr25_-_21716326 | 0.98 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr1_-_8101495 | 0.97 |
ENSDART00000161938
|
si:dkeyp-9d4.3
|
si:dkeyp-9d4.3 |
| chr2_+_24199276 | 0.96 |
ENSDART00000140575
|
map4l
|
microtubule associated protein 4 like |
| chr9_+_34334156 | 0.96 |
ENSDART00000144272
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr5_+_42467867 | 0.95 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr3_+_47245764 | 0.95 |
ENSDART00000193581
|
tnfsf14
|
TNF superfamily member 14 |
| chr10_+_439692 | 0.95 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr13_-_33009734 | 0.94 |
ENSDART00000134140
|
rbm25a
|
RNA binding motif protein 25a |
| chr8_+_36339470 | 0.94 |
ENSDART00000191250
|
FO744840.1
|
|
| chr22_+_9429710 | 0.94 |
ENSDART00000184089
|
si:ch211-11p18.6
|
si:ch211-11p18.6 |
| chr10_+_573667 | 0.93 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr22_+_1300587 | 0.93 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr12_+_10062953 | 0.92 |
ENSDART00000148689
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr4_+_77740228 | 0.92 |
ENSDART00000193397
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr19_+_9305964 | 0.91 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr24_-_18919562 | 0.91 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr13_+_27232694 | 0.90 |
ENSDART00000131128
|
rin2
|
Ras and Rab interactor 2 |
| chr11_+_1796426 | 0.90 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr7_-_34339845 | 0.89 |
ENSDART00000173816
|
madd
|
MAP-kinase activating death domain |
| chr10_-_32524035 | 0.88 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr5_-_32882162 | 0.87 |
ENSDART00000085769
|
lrsam1
|
leucine rich repeat and sterile alpha motif containing 1 |
| chr6_-_38930726 | 0.86 |
ENSDART00000154151
|
hdac7b
|
histone deacetylase 7b |
| chr12_+_47794089 | 0.85 |
ENSDART00000160726
|
polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A |
| chr17_-_50220228 | 0.84 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr23_-_20100436 | 0.84 |
ENSDART00000143445
|
si:dkey-32e6.6
|
si:dkey-32e6.6 |
| chr12_-_6880694 | 0.84 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr2_+_24199073 | 0.83 |
ENSDART00000144110
|
map4l
|
microtubule associated protein 4 like |
| chr24_-_1356668 | 0.83 |
ENSDART00000188935
|
nrp1a
|
neuropilin 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx15_tbx1_mgaa_tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.7 | 2.6 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.7 | 2.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.6 | 1.9 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.6 | 8.9 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.6 | 2.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.5 | 1.6 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.5 | 3.2 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.5 | 1.4 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.4 | 1.3 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.4 | 1.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.4 | 1.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.4 | 1.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 1.1 | GO:0034773 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.4 | 2.9 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.4 | 1.1 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.3 | 1.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 3.4 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.3 | 2.0 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.3 | 1.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.3 | 2.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.3 | 1.1 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.3 | 1.9 | GO:0019614 | catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.3 | 1.6 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.2 | 1.5 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.2 | 1.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 1.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 1.9 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 3.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 2.2 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.2 | 2.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 0.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 1.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.2 | 4.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.2 | 2.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.2 | 1.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 1.9 | GO:0019430 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.2 | 0.5 | GO:1902410 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.2 | 0.7 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.2 | 1.5 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.2 | 3.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.2 | 0.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.2 | 0.8 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.2 | 0.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.8 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 5.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.4 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.1 | 1.5 | GO:0035587 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.4 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.1 | 0.7 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 1.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.7 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 2.6 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 1.8 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 2.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 1.0 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.1 | 2.9 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.4 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.1 | 1.4 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.1 | 0.5 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.3 | GO:0002839 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 1.3 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.8 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0015824 | proline transport(GO:0015824) |
| 0.1 | 1.5 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 1.1 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 0.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 2.7 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.1 | 0.3 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.2 | GO:0051793 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.1 | 0.2 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.1 | 3.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.2 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.5 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.0 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.5 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 1.0 | GO:1901381 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.4 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 2.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 1.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 2.6 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 1.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.0 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.5 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 1.2 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.4 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 0.4 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.1 | 0.6 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.2 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.6 | GO:0050688 | regulation of response to biotic stimulus(GO:0002831) regulation of defense response to virus(GO:0050688) |
| 0.0 | 4.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 3.7 | GO:1902284 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 3.8 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 0.4 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.7 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0098581 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.6 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 3.7 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.7 | GO:0060078 | regulation of postsynaptic membrane potential(GO:0060078) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 3.0 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.4 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.5 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.2 | GO:0031444 | skeletal muscle thin filament assembly(GO:0030240) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 2.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.3 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 2.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 5.9 | GO:0034765 | regulation of ion transmembrane transport(GO:0034765) |
| 0.0 | 0.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 2.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.6 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 | 2.6 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 2.4 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.5 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.0 | 2.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.2 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.5 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.0 | 3.3 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.2 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.2 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.6 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 1.5 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 2.0 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.0 | 0.9 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.6 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0090303 | positive regulation of wound healing(GO:0090303) |
| 0.0 | 0.7 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.3 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.3 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.2 | GO:0033338 | medial fin development(GO:0033338) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0044550 | melanin biosynthetic process(GO:0042438) secondary metabolite biosynthetic process(GO:0044550) |
| 0.0 | 0.8 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.4 | 3.2 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.3 | 3.4 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.3 | 4.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.3 | 0.8 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 1.0 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 1.4 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.2 | 2.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.9 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 1.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 2.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.2 | 3.4 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.2 | 1.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 0.7 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.7 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 2.0 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.0 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 1.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.1 | 2.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 0.5 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 2.0 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 8.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 1.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 2.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 6.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.3 | GO:0036477 | somatodendritic compartment(GO:0036477) |
| 0.0 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 5.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 3.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 2.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 2.8 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 3.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0030666 | clathrin coat of endocytic vesicle(GO:0030128) endocytic vesicle membrane(GO:0030666) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 11.6 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.5 | 2.7 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.5 | 1.6 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.5 | 1.9 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 1.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.4 | 1.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.3 | 1.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 3.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.3 | 8.2 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 1.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.3 | 1.6 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.3 | 2.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.3 | 2.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.3 | 1.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.3 | 0.8 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 1.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 3.4 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.2 | 1.9 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.2 | 1.7 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.2 | 2.9 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.2 | 0.8 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 1.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 3.4 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.2 | 1.5 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.2 | 0.6 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.8 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.4 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 2.6 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 1.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.5 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.2 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.1 | 0.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 2.3 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 2.0 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 2.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 3.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 0.7 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.5 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.9 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 1.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 2.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.2 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.7 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 2.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 8.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.1 | 0.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 9.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.7 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.1 | 0.2 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.1 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 2.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 1.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 1.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.8 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 2.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 3.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 4.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 10.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.5 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.0 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.6 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 2.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0004155 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 4.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0004960 | thromboxane receptor activity(GO:0004960) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 6.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 1.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 4.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.2 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 13.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0099604 | ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 1.6 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.5 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.2 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 4.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.2 | 3.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 5.1 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 1.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.4 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.4 | 2.9 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 6.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 5.3 | REACTOME CA DEPENDENT EVENTS | Genes involved in Ca-dependent events |
| 0.2 | 2.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 3.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 2.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 1.9 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 1.7 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |
| 0.2 | 2.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 2.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 5.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.8 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.1 | 1.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 0.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 2.5 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 1.0 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.6 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 4.2 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 0.4 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.1 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.6 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.3 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.4 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |