Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
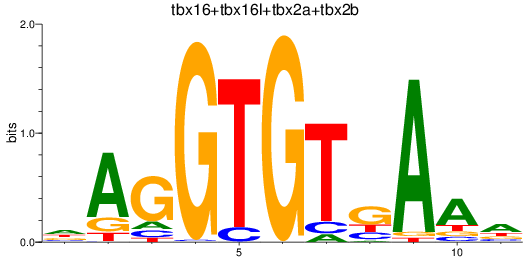
Results for tbr1b_tbx16+tbx16l+tbx2a+tbx2b
Z-value: 1.23
Transcription factors associated with tbr1b_tbx16+tbx16l+tbx2a+tbx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbr1b
|
ENSDARG00000004712 | T-box brain transcription factor 1b |
|
tbx2b
|
ENSDARG00000006120 | T-box transcription factor 2b |
|
tbx16l
|
ENSDARG00000006939 | T-box transcription factor 16, like |
|
tbx16
|
ENSDARG00000007329 | T-box transcription factor 16 |
|
tbx2a
|
ENSDARG00000018025 | T-box transcription factor 2a |
|
tbx2a
|
ENSDARG00000109541 | T-box transcription factor 2a |
|
tbx2b
|
ENSDARG00000116135 | T-box transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx6l | dr11_v1_chr5_+_42280372_42280372 | 0.95 | 6.5e-10 | Click! |
| tbx16 | dr11_v1_chr8_-_51753604_51753604 | 0.89 | 2.9e-07 | Click! |
| tbx2a | dr11_v1_chr5_-_56513825_56513825 | 0.59 | 7.9e-03 | Click! |
| tbr1b | dr11_v1_chr9_-_51436377_51436377 | -0.52 | 2.2e-02 | Click! |
| tbx2b | dr11_v1_chr15_+_27364394_27364394 | 0.03 | 9.1e-01 | Click! |
Activity profile of tbr1b_tbx16+tbx16l+tbx2a+tbx2b motif
Sorted Z-values of tbr1b_tbx16+tbx16l+tbx2a+tbx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16720196 | 5.03 |
ENSDART00000187639
|
si:dkey-26g8.4
|
si:dkey-26g8.4 |
| chr12_-_16619449 | 4.58 |
ENSDART00000182074
|
ctslb
|
cathepsin Lb |
| chr11_-_11575070 | 3.28 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr6_+_56141852 | 3.11 |
ENSDART00000149665
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr23_+_23232136 | 3.03 |
ENSDART00000126479
ENSDART00000187764 |
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr13_+_18533005 | 2.86 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr10_-_1961930 | 2.86 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr17_+_38255105 | 2.78 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr12_+_6002715 | 2.76 |
ENSDART00000114961
|
si:ch211-131k2.3
|
si:ch211-131k2.3 |
| chr9_+_15837398 | 2.75 |
ENSDART00000141063
|
si:dkey-103d23.5
|
si:dkey-103d23.5 |
| chr17_-_8312923 | 2.72 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr4_+_15006217 | 2.43 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr19_-_9776075 | 2.37 |
ENSDART00000140332
|
si:dkey-14o18.1
|
si:dkey-14o18.1 |
| chr3_-_32603191 | 2.36 |
ENSDART00000150997
|
si:ch73-248e21.7
|
si:ch73-248e21.7 |
| chr16_+_53203370 | 2.34 |
ENSDART00000154669
|
si:ch211-269k10.2
|
si:ch211-269k10.2 |
| chr13_-_50614639 | 2.22 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr20_+_34374735 | 2.18 |
ENSDART00000144090
|
si:dkeyp-11g8.3
|
si:dkeyp-11g8.3 |
| chr10_-_1961576 | 2.04 |
ENSDART00000042441
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr4_-_77216726 | 2.03 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr17_+_51517750 | 2.01 |
ENSDART00000180896
ENSDART00000193528 |
pxdn
|
peroxidasin |
| chr23_-_16734009 | 1.97 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr7_+_29167744 | 1.97 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr12_-_16636627 | 1.96 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr11_-_11791718 | 1.95 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr13_+_2894536 | 1.95 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr19_-_7420867 | 1.93 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr25_+_36292057 | 1.92 |
ENSDART00000152329
|
bmb
|
brambleberry |
| chr17_+_26965351 | 1.91 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr19_-_20148469 | 1.89 |
ENSDART00000134476
|
si:ch211-155k24.1
|
si:ch211-155k24.1 |
| chr20_+_48782068 | 1.86 |
ENSDART00000159275
|
nkx2.2b
|
NK2 homeobox 2b |
| chr14_+_97017 | 1.86 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr2_+_45081489 | 1.86 |
ENSDART00000123966
|
chrng
|
cholinergic receptor, nicotinic, gamma |
| chr5_-_32505109 | 1.86 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr3_-_34656745 | 1.83 |
ENSDART00000144824
|
znfl1
|
zinc finger-like gene 1 |
| chr25_+_418932 | 1.83 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr8_-_50287949 | 1.82 |
ENSDART00000023639
|
nkx2.7
|
NK2 transcription factor related 7 |
| chr21_+_39941184 | 1.73 |
ENSDART00000133604
|
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr7_-_38689562 | 1.72 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr9_-_7390388 | 1.70 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr9_-_31763246 | 1.70 |
ENSDART00000114742
|
si:dkey-250i3.3
|
si:dkey-250i3.3 |
| chr22_-_4439311 | 1.69 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr15_-_5580093 | 1.68 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr7_+_15329819 | 1.64 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr22_-_3152357 | 1.64 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr7_-_6279138 | 1.63 |
ENSDART00000173299
|
si:ch211-220f21.2
|
si:ch211-220f21.2 |
| chr8_-_11229523 | 1.63 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr2_-_14793343 | 1.63 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr4_+_54519511 | 1.62 |
ENSDART00000161653
|
znf974
|
zinc finger protein 974 |
| chr5_+_27525477 | 1.60 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr22_+_10678141 | 1.60 |
ENSDART00000193341
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr7_-_28696556 | 1.59 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr10_+_19039507 | 1.52 |
ENSDART00000193538
ENSDART00000111952 ENSDART00000180093 |
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr12_+_27231212 | 1.52 |
ENSDART00000133023
ENSDART00000123739 |
tmem106a
|
transmembrane protein 106A |
| chr19_-_31007417 | 1.52 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr14_-_2979878 | 1.51 |
ENSDART00000031211
|
bicc2
|
bicaudal C homolog 2 |
| chr3_+_43086548 | 1.49 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr9_+_34151367 | 1.45 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr2_-_42960353 | 1.42 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr7_+_53755054 | 1.40 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr3_+_31600593 | 1.39 |
ENSDART00000076640
ENSDART00000148189 |
ccdc43
|
coiled-coil domain containing 43 |
| chr24_-_34680956 | 1.39 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr23_-_24146591 | 1.39 |
ENSDART00000133269
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr9_+_30633184 | 1.39 |
ENSDART00000191310
|
tbc1d4
|
TBC1 domain family, member 4 |
| chr13_-_51922290 | 1.37 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr13_-_37127970 | 1.36 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr7_+_25221757 | 1.35 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr3_-_23643751 | 1.35 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr13_+_14976108 | 1.34 |
ENSDART00000011520
|
noto
|
notochord homeobox |
| chr17_+_34186632 | 1.33 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr18_+_9323211 | 1.33 |
ENSDART00000166114
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr23_+_42254960 | 1.32 |
ENSDART00000102980
|
zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr20_-_43723860 | 1.30 |
ENSDART00000122051
|
mixl1
|
Mix paired-like homeobox |
| chr2_+_15069011 | 1.30 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr7_-_48667056 | 1.29 |
ENSDART00000006378
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr7_+_15313443 | 1.29 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr16_-_32233463 | 1.28 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr13_-_36844945 | 1.27 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr11_-_25257045 | 1.27 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr2_-_6373829 | 1.26 |
ENSDART00000081633
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr8_-_48847772 | 1.24 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr1_+_26012668 | 1.23 |
ENSDART00000182428
|
BX323448.1
|
|
| chr13_-_35808904 | 1.21 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr15_-_36365840 | 1.20 |
ENSDART00000192926
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr2_+_35595454 | 1.17 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr12_-_35386910 | 1.17 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr4_-_56151174 | 1.16 |
ENSDART00000125904
|
znf986
|
zinc finger protein 986 |
| chr24_-_27419198 | 1.15 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr3_-_7464250 | 1.15 |
ENSDART00000159873
|
znf1001
|
zinc finger protein 1001 |
| chr4_+_64981411 | 1.14 |
ENSDART00000157798
|
CT955963.1
|
|
| chr8_+_3431671 | 1.14 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr14_-_43586150 | 1.13 |
ENSDART00000113697
|
eif4ea
|
eukaryotic translation initiation factor 4ea |
| chr9_+_54984900 | 1.11 |
ENSDART00000191622
|
mospd2
|
motile sperm domain containing 2 |
| chr10_-_13116337 | 1.10 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr6_-_8311044 | 1.10 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr12_-_4317165 | 1.09 |
ENSDART00000098041
|
zgc:153760
|
zgc:153760 |
| chr1_-_53277413 | 1.09 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr1_+_29183962 | 1.08 |
ENSDART00000113735
|
cars2
|
cysteinyl-tRNA synthetase 2, mitochondrial |
| chr24_-_18919562 | 1.07 |
ENSDART00000144244
ENSDART00000106188 ENSDART00000182518 |
cpa6
|
carboxypeptidase A6 |
| chr20_+_26394324 | 1.05 |
ENSDART00000078093
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr5_-_29512538 | 1.04 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr4_+_33461796 | 1.03 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr7_-_18508815 | 1.02 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr11_-_23219367 | 1.02 |
ENSDART00000003646
|
optc
|
opticin |
| chr24_+_26337623 | 1.02 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr8_+_12951155 | 1.02 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr13_+_29519115 | 1.02 |
ENSDART00000086711
|
chst3a
|
carbohydrate (chondroitin 6) sulfotransferase 3a |
| chr13_+_1381953 | 1.01 |
ENSDART00000019983
|
rab23
|
RAB23, member RAS oncogene family |
| chr11_-_3535537 | 1.01 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr13_+_45332203 | 1.01 |
ENSDART00000110466
|
si:ch211-168h21.3
|
si:ch211-168h21.3 |
| chr23_-_9991060 | 1.01 |
ENSDART00000111518
|
plxnb1a
|
plexin b1a |
| chr11_-_25257595 | 1.00 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr4_+_12031958 | 1.00 |
ENSDART00000044154
|
tnnt2c
|
troponin T2c, cardiac |
| chr19_-_18418763 | 0.99 |
ENSDART00000167271
|
zgc:112966
|
zgc:112966 |
| chr6_+_59840360 | 0.98 |
ENSDART00000154647
|
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr5_-_26198999 | 0.98 |
ENSDART00000188421
ENSDART00000183329 |
rad17
|
RAD17 checkpoint clamp loader component |
| chr6_-_21534301 | 0.98 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr17_-_43031763 | 0.98 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr7_+_51795667 | 0.98 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr13_+_15941850 | 0.97 |
ENSDART00000016294
|
fignl1
|
fidgetin-like 1 |
| chr5_-_69316142 | 0.97 |
ENSDART00000157238
ENSDART00000144570 |
smtnb
|
smoothelin b |
| chr15_-_23482088 | 0.97 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr25_-_27729046 | 0.96 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr6_+_8153813 | 0.95 |
ENSDART00000192436
|
nfil3-3
|
nuclear factor, interleukin 3 regulated, member 3 |
| chr4_+_32156734 | 0.95 |
ENSDART00000163904
|
CR450785.1
|
|
| chr5_-_39736383 | 0.94 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr19_-_1002959 | 0.94 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr7_-_41338923 | 0.94 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr14_+_48862987 | 0.93 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr23_-_29003864 | 0.93 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr23_+_44080382 | 0.93 |
ENSDART00000011715
|
zgc:56304
|
zgc:56304 |
| chr25_-_17579701 | 0.93 |
ENSDART00000073684
|
mmp15a
|
matrix metallopeptidase 15a |
| chr4_-_13973213 | 0.93 |
ENSDART00000067177
ENSDART00000144127 |
prickle1b
|
prickle homolog 1b |
| chr1_+_30723677 | 0.92 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr12_+_3912544 | 0.92 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr17_-_12758171 | 0.92 |
ENSDART00000131564
|
brms1la
|
breast cancer metastasis-suppressor 1-like a |
| chr1_+_34696503 | 0.92 |
ENSDART00000186106
|
CR339054.2
|
|
| chr24_+_22485710 | 0.91 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr25_-_11026907 | 0.91 |
ENSDART00000156846
|
mespbb
|
mesoderm posterior bb |
| chr1_-_55888970 | 0.91 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr13_+_27232848 | 0.90 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr14_+_45406299 | 0.90 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr10_+_26944418 | 0.90 |
ENSDART00000135493
|
frmd8
|
FERM domain containing 8 |
| chr4_-_60049792 | 0.90 |
ENSDART00000158199
|
znf1033
|
zinc finger protein 1033 |
| chr15_-_47848544 | 0.89 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr6_-_40651944 | 0.89 |
ENSDART00000187423
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr8_+_23245596 | 0.89 |
ENSDART00000143313
|
si:ch211-196c10.11
|
si:ch211-196c10.11 |
| chr14_-_42231293 | 0.89 |
ENSDART00000185486
|
BX890543.1
|
|
| chr25_+_25085349 | 0.88 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr25_-_26736088 | 0.88 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr6_-_53281518 | 0.88 |
ENSDART00000157621
|
rbm5
|
RNA binding motif protein 5 |
| chr7_+_39166460 | 0.88 |
ENSDART00000052318
ENSDART00000146635 ENSDART00000173877 ENSDART00000173767 ENSDART00000173600 |
mdka
|
midkine a |
| chr17_-_8907668 | 0.88 |
ENSDART00000155996
ENSDART00000155624 |
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr16_+_14707960 | 0.87 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr2_-_39675829 | 0.86 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr22_-_34872533 | 0.86 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr4_+_17642731 | 0.86 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr1_+_30723380 | 0.85 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr25_+_7435291 | 0.85 |
ENSDART00000172567
ENSDART00000163017 |
prc1a
|
protein regulator of cytokinesis 1a |
| chr16_-_7379328 | 0.84 |
ENSDART00000060447
|
med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr3_-_53533128 | 0.84 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr11_+_24851671 | 0.84 |
ENSDART00000167659
|
ipo9
|
importin 9 |
| chr7_+_20917966 | 0.84 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr18_-_10713414 | 0.84 |
ENSDART00000034817
|
rbm28
|
RNA binding motif protein 28 |
| chr21_+_39941559 | 0.83 |
ENSDART00000189718
ENSDART00000160875 ENSDART00000135235 |
slc47a1
|
solute carrier family 47 (multidrug and toxin extrusion), member 1 |
| chr13_+_28495419 | 0.83 |
ENSDART00000025583
|
fgf8a
|
fibroblast growth factor 8a |
| chr13_+_21677767 | 0.83 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr22_+_2032023 | 0.82 |
ENSDART00000171662
|
znf1174
|
zinc finger protein 1174 |
| chr11_+_25257022 | 0.82 |
ENSDART00000156052
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr2_-_7185460 | 0.82 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr21_-_929448 | 0.82 |
ENSDART00000133976
|
txnl1
|
thioredoxin-like 1 |
| chr24_+_21174851 | 0.82 |
ENSDART00000154940
ENSDART00000155977 ENSDART00000122762 |
naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_-_52505279 | 0.82 |
ENSDART00000052907
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr2_+_327081 | 0.81 |
ENSDART00000155595
|
zgc:174263
|
zgc:174263 |
| chr11_+_42494531 | 0.80 |
ENSDART00000067604
|
arf4a
|
ADP-ribosylation factor 4a |
| chr13_+_45967179 | 0.80 |
ENSDART00000074499
|
olig4
|
oligodendrocyte transcription factor 4 |
| chr17_+_23494698 | 0.80 |
ENSDART00000155150
|
kif20ba
|
kinesin family member 20Ba |
| chr4_+_14981854 | 0.79 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr1_-_51219965 | 0.79 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr4_-_11580948 | 0.79 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr24_+_9693951 | 0.78 |
ENSDART00000082411
|
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr21_-_5831413 | 0.77 |
ENSDART00000150914
|
wu:fj64h06
|
wu:fj64h06 |
| chr22_-_34609528 | 0.77 |
ENSDART00000190781
ENSDART00000171712 |
terf2ip
|
telomeric repeat binding factor 2, interacting protein |
| chr19_-_3878548 | 0.77 |
ENSDART00000168377
ENSDART00000172271 |
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr22_+_25734180 | 0.77 |
ENSDART00000143367
|
si:dkeyp-98a7.9
|
si:dkeyp-98a7.9 |
| chr16_+_15114645 | 0.77 |
ENSDART00000158483
|
mtbp
|
MDM2 binding protein |
| chr10_-_41664427 | 0.76 |
ENSDART00000150213
|
ggt1b
|
gamma-glutamyltransferase 1b |
| chr21_+_17956856 | 0.76 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr21_-_19919020 | 0.76 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr14_+_28518349 | 0.76 |
ENSDART00000159961
|
stag2b
|
stromal antigen 2b |
| chr13_+_15702142 | 0.75 |
ENSDART00000135960
|
trmt61a
|
tRNA methyltransferase 61A |
| chr21_+_26726936 | 0.75 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr2_-_17393216 | 0.75 |
ENSDART00000123137
|
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr20_-_31238313 | 0.75 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr1_-_52498146 | 0.75 |
ENSDART00000122217
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr14_-_32744464 | 0.75 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr7_-_48665305 | 0.74 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr2_-_17392799 | 0.73 |
ENSDART00000136470
ENSDART00000141188 |
st3gal3b
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3b |
| chr22_+_15959844 | 0.73 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr1_+_8521323 | 0.73 |
ENSDART00000121439
ENSDART00000103626 ENSDART00000141283 |
mief2
|
mitochondrial elongation factor 2 |
| chr25_-_11016675 | 0.73 |
ENSDART00000099572
|
mespab
|
mesoderm posterior ab |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbr1b_tbx16+tbx16l+tbx2a+tbx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.9 | 5.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.8 | 3.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.6 | 1.9 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.5 | 3.7 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 2.2 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.4 | 0.8 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.4 | 1.4 | GO:0048327 | axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 0.3 | 0.3 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.3 | 1.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 1.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 3.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.3 | 1.2 | GO:0070199 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 2.0 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 0.3 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.3 | 0.8 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.3 | 1.4 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.3 | 0.8 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.3 | 1.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 1.4 | GO:0021855 | hypothalamus cell migration(GO:0021855) |
| 0.3 | 1.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.3 | 1.6 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.3 | 1.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 0.8 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 3.1 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 1.4 | GO:0061193 | tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) |
| 0.2 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.9 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.2 | 0.7 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.2 | 0.7 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 0.7 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.6 | GO:0060765 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 | 0.6 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 0.6 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.2 | 1.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 2.5 | GO:0003207 | cardiac chamber formation(GO:0003207) |
| 0.2 | 1.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.1 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.2 | 1.9 | GO:0021508 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.2 | 0.9 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.2 | 2.7 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 0.9 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.2 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.9 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.2 | 1.7 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 1.9 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 0.8 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 2.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.2 | 0.5 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 1.0 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.2 | 0.5 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 0.9 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.2 | 4.6 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 0.7 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 1.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.6 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 2.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.3 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.1 | 2.3 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.8 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) glial cell fate specification(GO:0021780) |
| 0.1 | 0.4 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 0.4 | GO:2000257 | regulation of protein activation cascade(GO:2000257) |
| 0.1 | 2.2 | GO:0003128 | heart field specification(GO:0003128) |
| 0.1 | 0.8 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 1.5 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 1.8 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 0.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.2 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 1.0 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.1 | 0.9 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 0.5 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.7 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 1.3 | GO:1990399 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 1.4 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.7 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 2.5 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 1.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 0.4 | GO:0009202 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) deoxyribonucleoside triphosphate biosynthetic process(GO:0009202) dGDP metabolic process(GO:0046066) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.7 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 0.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 0.4 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.6 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.4 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.1 | 0.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.5 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.9 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.1 | 0.8 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.8 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.4 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 2.0 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.7 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.8 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 0.2 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.1 | 0.5 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.6 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.6 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.1 | 0.3 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) |
| 0.1 | 0.8 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.3 | GO:0043102 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.1 | 0.5 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 1.3 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.5 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.0 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 2.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 1.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.4 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.2 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.9 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 0.3 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.1 | 0.2 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.1 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.1 | 2.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 2.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.2 | GO:0006953 | acute inflammatory response(GO:0002526) acute-phase response(GO:0006953) |
| 0.1 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.2 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 1.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.8 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.3 | GO:1903076 | regulation of protein localization to plasma membrane(GO:1903076) regulation of protein localization to cell periphery(GO:1904375) |
| 0.0 | 1.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.4 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 2.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.4 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:1903722 | regulation of centriole elongation(GO:1903722) negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.0 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.3 | GO:0050779 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.0 | 0.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0034205 | Notch receptor processing(GO:0007220) beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 1.2 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.3 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 3.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:0045687 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.7 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.9 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 1.6 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.8 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 1.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.7 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.3 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 1.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) xanthophore differentiation(GO:0050936) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.5 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.3 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.7 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.3 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 1.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.3 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 1.0 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.5 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.7 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.3 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 1.7 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.8 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0002423 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) interferon-gamma production(GO:0032609) regulation of interferon-gamma production(GO:0032649) positive regulation of interferon-gamma production(GO:0032729) positive regulation of mast cell activation(GO:0033005) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.2 | GO:0008343 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 1.8 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0046634 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) negative regulation of cell fate commitment(GO:0010454) regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043370) negative regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043371) negative regulation of T cell differentiation(GO:0045581) regulation of T-helper cell differentiation(GO:0045622) negative regulation of T-helper cell differentiation(GO:0045623) regulation of alpha-beta T cell activation(GO:0046634) negative regulation of alpha-beta T cell activation(GO:0046636) regulation of alpha-beta T cell differentiation(GO:0046637) negative regulation of alpha-beta T cell differentiation(GO:0046639) regulation of T-helper 17 type immune response(GO:2000316) negative regulation of T-helper 17 type immune response(GO:2000317) regulation of T-helper 17 cell differentiation(GO:2000319) negative regulation of T-helper 17 cell differentiation(GO:2000320) regulation of T-helper 17 cell lineage commitment(GO:2000328) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) regulation of CD4-positive, alpha-beta T cell activation(GO:2000514) negative regulation of CD4-positive, alpha-beta T cell activation(GO:2000515) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.4 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0060043 | regulation of organ growth(GO:0046620) regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) regulation of heart growth(GO:0060420) |
| 0.0 | 0.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 3.0 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.9 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.5 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.0 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.4 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 1.5 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 1.5 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.0 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 1.2 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.3 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 10.0 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.4 | GO:0021537 | telencephalon development(GO:0021537) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 2.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 1.4 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.5 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.6 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.8 | GO:0044744 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 1.4 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.4 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.2 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.5 | GO:0034204 | lipid translocation(GO:0034204) |
| 0.0 | 0.2 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 1.3 | GO:0030902 | hindbrain development(GO:0030902) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) negative regulation of sodium ion transport(GO:0010766) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.4 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.0 | 4.7 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.5 | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage(GO:0008630) |
| 0.0 | 0.3 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.4 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 0.1 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.4 | GO:0006986 | response to unfolded protein(GO:0006986) response to topologically incorrect protein(GO:0035966) |
| 0.0 | 0.2 | GO:0033338 | medial fin development(GO:0033338) |
| 0.0 | 0.4 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:0032835 | glomerulus development(GO:0032835) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.7 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 1.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.2 | 1.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.2 | 0.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 2.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.2 | 0.8 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 1.3 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.5 | GO:0031511 | condensed chromosome inner kinetochore(GO:0000939) Mis6-Sim4 complex(GO:0031511) |
| 0.1 | 1.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 1.0 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.6 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 0.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.5 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.3 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.1 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.4 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.1 | 0.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 2.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.7 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.5 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.1 | 0.8 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 2.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.6 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.4 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0044545 | NSL complex(GO:0044545) |
| 0.1 | 1.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.5 | GO:0000796 | condensin complex(GO:0000796) |
| 0.1 | 1.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 0.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.1 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 0.2 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 1.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.4 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.5 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.9 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.6 | GO:0071004 | U2-type prespliceosome(GO:0071004) prespliceosome(GO:0071010) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 8.2 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 4.9 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 0.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.9 | GO:0030532 | small nuclear ribonucleoprotein complex(GO:0030532) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.0 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 1.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 3.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:1990752 | microtubule end(GO:1990752) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0097223 | sperm flagellum(GO:0036126) sperm part(GO:0097223) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 1.9 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.6 | 2.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.4 | 1.6 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.4 | 5.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.4 | 1.9 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 1.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.3 | 1.7 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.3 | 1.0 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 4.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.3 | 1.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.3 | 1.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 0.8 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 1.0 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.3 | 2.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 2.4 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.2 | 0.8 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 0.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.2 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 1.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 2.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.2 | 0.9 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 0.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 0.5 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.2 | 0.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 1.3 | GO:0008026 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.2 | 1.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 2.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.7 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 0.7 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.1 | 2.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.1 | 0.6 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.1 | 1.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 1.4 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 0.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 2.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.7 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.7 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.3 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.4 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 1.0 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 1.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.0 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.9 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.3 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.4 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.4 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.4 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.2 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.1 | 1.7 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 0.4 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.6 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.3 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.7 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.7 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 2.0 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 2.8 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 1.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 0.6 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.0 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.5 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.4 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 1.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.4 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.6 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.9 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 1.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0015462 | protein-transmembrane transporting ATPase activity(GO:0015462) |
| 0.0 | 0.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.3 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.8 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 2.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 4.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 4.4 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 1.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 43.2 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.8 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.0 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.6 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.0 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 1.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.8 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.9 | GO:0019901 | protein kinase binding(GO:0019901) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 5.6 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 0.8 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.4 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.7 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.8 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 0.7 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.1 | 2.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 0.8 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.1 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.5 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID MTOR 4PATHWAY | mTOR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 1.9 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.0 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.5 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 2.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 3.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 0.7 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.9 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 1.0 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.1 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.0 | 0.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 1.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.5 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.3 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |