Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
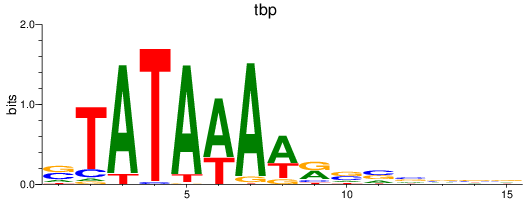
Results for tbp
Z-value: 6.41
Transcription factors associated with tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbp
|
ENSDARG00000014994 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbp | dr11_v1_chr13_-_24396199_24396199 | -0.74 | 3.0e-04 | Click! |
Activity profile of tbp motif
Sorted Z-values of tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22240052 | 56.19 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr9_-_22232902 | 52.07 |
ENSDART00000101845
|
crygm2d5
|
crystallin, gamma M2d5 |
| chr6_+_41181869 | 46.66 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr9_-_22245572 | 44.25 |
ENSDART00000114943
|
crygm2d4
|
crystallin, gamma M2d4 |
| chr9_-_22299412 | 38.32 |
ENSDART00000139101
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr9_-_22158784 | 37.47 |
ENSDART00000167850
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr9_-_22213297 | 36.43 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr9_-_22135576 | 35.00 |
ENSDART00000101902
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr16_-_25085327 | 34.16 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr6_-_346084 | 33.93 |
ENSDART00000162599
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr9_-_22281854 | 33.46 |
ENSDART00000146319
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr9_-_22318511 | 33.17 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr11_+_25477643 | 33.17 |
ENSDART00000065941
|
opn1lw1
|
opsin 1 (cone pigments), long-wave-sensitive, 1 |
| chr5_+_37837245 | 32.93 |
ENSDART00000171617
|
epd
|
ependymin |
| chr9_-_22117430 | 32.51 |
ENSDART00000101916
|
crygm2d15
|
crystallin, gamma M2d15 |
| chr9_-_22129788 | 32.12 |
ENSDART00000124272
ENSDART00000175417 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr9_-_22135420 | 31.77 |
ENSDART00000184959
|
crygm2d8
|
crystallin, gamma M2d8 |
| chr9_-_22272181 | 30.72 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr24_-_41312459 | 30.56 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr9_-_22158325 | 28.05 |
ENSDART00000114568
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr5_-_30615901 | 27.08 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr9_-_22182396 | 26.66 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr7_+_35068036 | 24.28 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr12_-_17712393 | 23.80 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr9_-_22188117 | 23.55 |
ENSDART00000132890
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr21_+_30043054 | 23.22 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr9_-_22339582 | 22.16 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr2_-_38000276 | 22.09 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr11_+_24002503 | 20.85 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr24_+_3307857 | 19.73 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr11_+_24001993 | 19.62 |
ENSDART00000168215
|
chia.2
|
chitinase, acidic.2 |
| chr10_-_44027391 | 19.51 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr9_-_22147567 | 19.49 |
ENSDART00000110941
|
crygm2d14
|
crystallin, gamma M2d14 |
| chr19_-_3193912 | 19.27 |
ENSDART00000133159
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr9_+_22080122 | 18.99 |
ENSDART00000065956
ENSDART00000136014 |
crygm2e
|
crystallin, gamma M2e |
| chr25_+_8356707 | 18.75 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr11_+_25472758 | 18.75 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr15_+_19682013 | 17.48 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr6_-_40755909 | 17.30 |
ENSDART00000026257
|
myhb
|
myosin, heavy chain b |
| chr17_+_27456804 | 16.65 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr8_+_48613040 | 16.01 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr5_-_33259079 | 15.78 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr19_-_3193443 | 15.46 |
ENSDART00000179855
|
si:ch211-133n4.6
|
si:ch211-133n4.6 |
| chr15_+_45643787 | 15.29 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr15_-_5815006 | 15.21 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr6_-_40756138 | 15.17 |
ENSDART00000156295
|
myhb
|
myosin, heavy chain b |
| chr21_+_25226558 | 15.13 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr3_+_33340939 | 14.71 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr12_-_35988586 | 14.53 |
ENSDART00000157746
|
pde6gb
|
phosphodiesterase 6G, cGMP-specific, rod, gamma, paralog b |
| chr5_-_71722257 | 14.18 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr15_+_15390882 | 13.79 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr10_-_17745345 | 13.59 |
ENSDART00000132690
ENSDART00000135376 |
si:dkey-200l5.4
|
si:dkey-200l5.4 |
| chr6_+_41186320 | 13.32 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr19_-_5345930 | 12.99 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr2_-_1443692 | 12.46 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr23_+_45579497 | 12.26 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr22_-_263117 | 11.62 |
ENSDART00000158134
|
zgc:66156
|
zgc:66156 |
| chr22_-_282498 | 11.53 |
ENSDART00000182766
|
CABZ01079178.1
|
|
| chr5_+_9348284 | 11.30 |
ENSDART00000149417
|
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr5_-_27867657 | 11.18 |
ENSDART00000112495
|
tcima
|
transcriptional and immune response regulator a |
| chr22_-_11626014 | 11.16 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr21_+_25216397 | 11.05 |
ENSDART00000130970
|
sycn.3
|
syncollin, tandem duplicate 3 |
| chr25_-_18454016 | 10.96 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr1_-_22338521 | 10.92 |
ENSDART00000176849
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr10_-_24371312 | 10.81 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr10_+_17026870 | 10.80 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr3_-_32818607 | 10.64 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr20_-_39273987 | 10.53 |
ENSDART00000127173
|
clu
|
clusterin |
| chr1_+_55137943 | 10.22 |
ENSDART00000138070
ENSDART00000150510 ENSDART00000133472 ENSDART00000136378 |
mb
|
myoglobin |
| chr21_-_28523548 | 10.08 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr11_+_3575963 | 9.99 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr9_+_22017368 | 9.54 |
ENSDART00000023059
|
zgc:153846
|
zgc:153846 |
| chr5_-_36837846 | 9.14 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr19_+_19976990 | 9.12 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr7_-_7823662 | 9.12 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr18_-_5527050 | 8.97 |
ENSDART00000145400
ENSDART00000132498 ENSDART00000146209 |
zgc:153317
|
zgc:153317 |
| chr7_-_16562200 | 8.90 |
ENSDART00000169093
ENSDART00000173491 |
csrp3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr15_+_19681718 | 8.87 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr3_+_59051503 | 8.84 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr6_+_39499623 | 8.84 |
ENSDART00000036057
|
si:ch211-173n18.3
|
si:ch211-173n18.3 |
| chr25_+_8407892 | 8.81 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr7_-_73851280 | 8.81 |
ENSDART00000190053
|
FP236812.3
|
|
| chr3_-_57425961 | 8.72 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr14_-_33872092 | 8.72 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr14_-_11507211 | 8.70 |
ENSDART00000186873
ENSDART00000109181 ENSDART00000186166 ENSDART00000186986 |
zgc:174917
|
zgc:174917 |
| chr6_+_3934738 | 8.67 |
ENSDART00000159673
|
dync1i2b
|
dynein, cytoplasmic 1, intermediate chain 2b |
| chr23_-_17429775 | 8.66 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr9_+_1139378 | 8.65 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr10_-_39321367 | 8.63 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr25_+_30298377 | 8.55 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr1_+_29096881 | 8.45 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr10_-_22845485 | 8.34 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr22_+_18530395 | 8.21 |
ENSDART00000105415
ENSDART00000183958 |
si:ch211-212d10.1
|
si:ch211-212d10.1 |
| chr4_+_17336557 | 8.07 |
ENSDART00000111650
|
pmch
|
pro-melanin-concentrating hormone |
| chr2_+_15100742 | 8.05 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr17_+_9308425 | 8.04 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr16_-_44349845 | 7.96 |
ENSDART00000170932
|
rims2a
|
regulating synaptic membrane exocytosis 2a |
| chr3_+_26135502 | 7.81 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr8_+_48609521 | 7.73 |
ENSDART00000060765
|
nppb
|
natriuretic peptide B |
| chr13_-_2215213 | 7.70 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr12_-_3903886 | 7.61 |
ENSDART00000184214
ENSDART00000041082 |
gdpd3b
|
glycerophosphodiester phosphodiesterase domain containing 3b |
| chr10_-_5058823 | 7.60 |
ENSDART00000139825
|
tmem150c
|
transmembrane protein 150C |
| chr20_+_27020201 | 7.57 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
| chr21_-_25685739 | 7.41 |
ENSDART00000129619
ENSDART00000101205 |
phkg1b
|
phosphorylase kinase, gamma 1b (muscle) |
| chr1_+_1599979 | 7.39 |
ENSDART00000097626
|
urp2
|
urotensin II-related peptide |
| chr10_+_17776981 | 7.38 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr23_-_35082494 | 7.33 |
ENSDART00000189809
|
BX294434.1
|
|
| chr18_+_14342326 | 7.22 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr14_-_2206476 | 7.04 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr3_+_15505275 | 6.75 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr11_+_34235372 | 6.72 |
ENSDART00000063150
|
fam43a
|
family with sequence similarity 43, member A |
| chr11_+_42478184 | 6.57 |
ENSDART00000089963
|
zgc:110286
|
zgc:110286 |
| chr3_-_59472422 | 6.52 |
ENSDART00000100327
|
nptx1l
|
neuronal pentraxin 1 like |
| chr24_+_37688729 | 6.47 |
ENSDART00000137017
|
h3f3d
|
H3 histone, family 3D |
| chr8_-_6877390 | 6.47 |
ENSDART00000170883
ENSDART00000005321 |
neflb
|
neurofilament, light polypeptide b |
| chr22_+_5752257 | 6.30 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr21_+_7823146 | 6.25 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr9_-_35334642 | 6.23 |
ENSDART00000157195
|
ncam2
|
neural cell adhesion molecule 2 |
| chr11_-_45152702 | 6.04 |
ENSDART00000168066
|
afmid
|
arylformamidase |
| chr22_-_294700 | 6.03 |
ENSDART00000189179
|
CABZ01079179.1
|
|
| chr14_-_11430566 | 5.91 |
ENSDART00000137154
ENSDART00000091158 |
irg1l
|
immunoresponsive gene 1, like |
| chr17_-_37214196 | 5.85 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr16_+_20161805 | 5.80 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr2_-_6482240 | 5.75 |
ENSDART00000132623
|
rgs13
|
regulator of G protein signaling 13 |
| chr14_-_46442459 | 5.75 |
ENSDART00000158770
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr10_-_43964028 | 5.73 |
ENSDART00000009134
ENSDART00000133450 |
sept5b
|
septin 5b |
| chr13_+_38520927 | 5.72 |
ENSDART00000137299
ENSDART00000111080 |
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr11_+_43403525 | 5.71 |
ENSDART00000180683
|
vipb
|
vasoactive intestinal peptide b |
| chr12_+_39034541 | 5.64 |
ENSDART00000182502
ENSDART00000169813 |
si:dkey-239b22.2
|
si:dkey-239b22.2 |
| chr5_-_32274383 | 5.51 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr20_+_42668875 | 5.47 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr24_-_5786759 | 5.37 |
ENSDART00000152069
|
chst2b
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2b |
| chr4_-_4780667 | 5.35 |
ENSDART00000133973
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr10_+_38593645 | 5.35 |
ENSDART00000011573
|
mmp13a
|
matrix metallopeptidase 13a |
| chr2_+_24507597 | 5.28 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr3_-_36606884 | 5.28 |
ENSDART00000172003
|
si:dkeyp-72e1.6
|
si:dkeyp-72e1.6 |
| chr3_+_49074008 | 5.18 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr7_-_66137065 | 5.17 |
ENSDART00000145843
|
pth1b
|
parathyroid hormone 1b |
| chr11_-_24016761 | 5.16 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr6_+_11249706 | 5.07 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr11_-_40030139 | 5.06 |
ENSDART00000021916
|
uts2b
|
urotensin 2, beta |
| chr4_+_842010 | 5.06 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr9_-_22355391 | 5.03 |
ENSDART00000009115
|
crygm3
|
crystallin, gamma M3 |
| chr15_-_44601331 | 4.99 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr6_+_11250033 | 4.99 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr7_+_29951997 | 4.97 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr13_-_20381485 | 4.89 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr6_+_39493864 | 4.87 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr14_-_2036604 | 4.83 |
ENSDART00000192446
|
BX005294.2
|
|
| chr5_-_13685047 | 4.75 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr4_+_1146346 | 4.70 |
ENSDART00000166467
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr21_+_25221940 | 4.64 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr22_+_7480465 | 4.62 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr6_-_54180699 | 4.61 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr23_+_27675581 | 4.55 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr12_-_29624638 | 4.54 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr9_-_52386733 | 4.53 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr23_-_35347714 | 4.50 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr24_+_37598441 | 4.49 |
ENSDART00000125145
|
rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila) |
| chr12_+_22607761 | 4.42 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr8_-_31369161 | 4.42 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr3_-_60401826 | 4.32 |
ENSDART00000144030
ENSDART00000160821 |
si:ch211-214b16.4
|
si:ch211-214b16.4 |
| chr5_+_4366431 | 4.14 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr2_+_24507770 | 4.13 |
ENSDART00000154802
ENSDART00000052063 |
rps28
|
ribosomal protein S28 |
| chr9_+_31075896 | 4.13 |
ENSDART00000188042
|
clybl
|
citrate lyase beta like |
| chr13_+_23176330 | 4.05 |
ENSDART00000168351
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr17_+_14711765 | 3.98 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr4_-_4780510 | 3.95 |
ENSDART00000109609
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr7_+_5906327 | 3.94 |
ENSDART00000173160
|
zgc:112234
|
zgc:112234 |
| chr2_-_53500424 | 3.94 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr5_+_21181047 | 3.91 |
ENSDART00000088506
|
arhgap25
|
Rho GTPase activating protein 25 |
| chr17_-_29312506 | 3.86 |
ENSDART00000133668
|
tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr20_-_36393555 | 3.77 |
ENSDART00000153421
|
si:dkey-1j5.4
|
si:dkey-1j5.4 |
| chr22_-_21392748 | 3.61 |
ENSDART00000144648
|
ankrd24
|
ankyrin repeat domain 24 |
| chr5_-_26181863 | 3.60 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr17_-_50234004 | 3.60 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr13_+_35745572 | 3.59 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr15_-_4967302 | 3.59 |
ENSDART00000101992
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr10_+_6010570 | 3.58 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr24_+_26402110 | 3.54 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr17_-_35361322 | 3.50 |
ENSDART00000019617
|
rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr18_+_17534627 | 3.48 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr2_+_35603637 | 3.47 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr2_-_7431590 | 3.39 |
ENSDART00000185699
|
asip2b
|
agouti signaling protein, nonagouti homolog (mouse) 2b |
| chr2_-_42091926 | 3.39 |
ENSDART00000142663
|
si:dkey-97a13.12
|
si:dkey-97a13.12 |
| chr3_+_5575313 | 3.39 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr15_+_1134870 | 3.36 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr2_-_30182353 | 3.28 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr9_+_42269059 | 3.23 |
ENSDART00000113435
|
si:dkey-10c21.1
|
si:dkey-10c21.1 |
| chr24_-_31425799 | 3.17 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr6_+_11250316 | 3.14 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr4_-_20485508 | 3.13 |
ENSDART00000183276
ENSDART00000023337 |
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr10_+_44924684 | 3.04 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
| chr5_-_60885935 | 2.96 |
ENSDART00000128350
|
rad51d
|
RAD51 paralog D |
| chr20_+_35438300 | 2.95 |
ENSDART00000102504
ENSDART00000153249 |
tdrd6
|
tudor domain containing 6 |
| chr10_-_24318538 | 2.94 |
ENSDART00000109549
ENSDART00000190656 |
inpp5kb
|
inositol polyphosphate-5-phosphatase Kb |
| chr19_+_32257472 | 2.92 |
ENSDART00000186471
|
atxn1a
|
ataxin 1a |
| chr10_+_42374957 | 2.85 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr6_-_23619428 | 2.82 |
ENSDART00000053126
|
aanat1
|
arylalkylamine N-acetyltransferase 1 |
| chr4_+_75314247 | 2.81 |
ENSDART00000162365
|
CABZ01041913.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.1 | 48.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 7.9 | 23.7 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 4.9 | 651.8 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 4.7 | 18.8 | GO:0044060 | regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) |
| 4.1 | 45.6 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 3.8 | 7.6 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 3.1 | 12.5 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 2.3 | 111.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 2.2 | 8.9 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 2.0 | 14.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 2.0 | 7.8 | GO:0031448 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 1.8 | 5.5 | GO:0015695 | organic cation transport(GO:0015695) |
| 1.4 | 5.7 | GO:0007620 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 1.3 | 13.2 | GO:0044805 | late nucleophagy(GO:0044805) |
| 1.3 | 3.9 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 1.3 | 15.3 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 1.2 | 8.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.2 | 3.6 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 1.2 | 3.6 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 1.2 | 4.7 | GO:0010459 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 1.1 | 5.7 | GO:0016322 | neuron remodeling(GO:0016322) |
| 1.1 | 8.7 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 1.1 | 8.5 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 1.0 | 4.1 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 1.0 | 8.7 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.9 | 3.5 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.8 | 10.8 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.8 | 8.6 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.8 | 3.1 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.8 | 6.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.7 | 3.0 | GO:0042148 | strand invasion(GO:0042148) |
| 0.7 | 5.9 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.7 | 31.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.6 | 10.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.6 | 14.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.6 | 2.4 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.6 | 8.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.5 | 10.9 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.5 | 10.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.4 | 4.2 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.4 | 2.4 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.4 | 1.9 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.4 | 11.2 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.4 | 10.0 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.4 | 8.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.4 | 2.5 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.3 | 0.3 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.3 | 5.4 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.3 | 11.2 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.3 | 2.9 | GO:0030719 | P granule organization(GO:0030719) |
| 0.3 | 3.5 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.3 | 38.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.3 | 3.6 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.3 | 1.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.3 | 5.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.3 | 1.0 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.3 | 1.0 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.2 | 1.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 | 1.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 2.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.2 | 5.9 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 5.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.2 | 11.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.2 | 3.9 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.2 | 1.0 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 4.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.2 | 1.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 0.3 | GO:1901533 | negative regulation of hematopoietic progenitor cell differentiation(GO:1901533) |
| 0.2 | 1.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 9.3 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.2 | 11.9 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.2 | 5.7 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 9.9 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.2 | 1.9 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.1 | 0.7 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 37.6 | GO:0006887 | exocytosis(GO:0006887) |
| 0.1 | 0.5 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 8.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 2.0 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 5.6 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.1 | 0.6 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 1.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 1.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 6.5 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.1 | 2.5 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.8 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 1.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 8.7 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) |
| 0.1 | 8.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 0.7 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 7.6 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.1 | 1.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.0 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 4.2 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 0.4 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 2.4 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.8 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 3.3 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 4.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 8.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.8 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 2.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 2.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 31.3 | GO:0006508 | proteolysis(GO:0006508) |
| 0.0 | 1.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.0 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 1.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 48.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 3.3 | 127.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 3.1 | 9.4 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 2.0 | 7.8 | GO:0031673 | H zone(GO:0031673) |
| 1.8 | 18.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 1.7 | 30.8 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 1.6 | 8.1 | GO:0005883 | neurofilament(GO:0005883) |
| 1.0 | 2.9 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.0 | 5.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.7 | 10.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.6 | 8.7 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.6 | 7.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.6 | 3.0 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.6 | 3.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.5 | 13.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.4 | 50.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 8.0 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.2 | 3.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 63.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.2 | 8.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 13.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 5.7 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 7.7 | GO:0016605 | PML body(GO:0016605) |
| 0.2 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 10.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 2.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 1.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 7.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 5.7 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.4 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 155.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 81.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 1.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.7 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.1 | 2.5 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 5.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 10.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 8.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 0.6 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 4.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 4.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 4.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.4 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.4 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 4.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 10.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.0 | 661.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 4.1 | 45.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 3.9 | 23.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 3.7 | 48.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 3.6 | 10.9 | GO:0042165 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 3.1 | 12.5 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 3.1 | 15.4 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 2.9 | 14.7 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 2.9 | 8.6 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 2.8 | 8.5 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 2.3 | 111.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 2.3 | 9.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 1.9 | 7.7 | GO:0005521 | lamin binding(GO:0005521) |
| 1.5 | 10.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 1.5 | 7.4 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 1.4 | 4.1 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 1.2 | 8.7 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 1.2 | 3.6 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 1.2 | 3.6 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 1.1 | 4.5 | GO:0070513 | death domain binding(GO:0070513) |
| 1.1 | 14.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.9 | 4.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.8 | 10.8 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.7 | 8.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.6 | 1.9 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.6 | 5.4 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.6 | 4.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.6 | 2.4 | GO:0032896 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.5 | 13.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.5 | 2.9 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.5 | 10.2 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.5 | 7.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.5 | 5.5 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.4 | 11.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 61.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.4 | 4.7 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.4 | 1.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 11.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.4 | 2.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.4 | 10.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 7.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.4 | 10.1 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.3 | 2.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.3 | 8.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.3 | 3.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 70.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 3.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.3 | 2.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 8.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 6.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 3.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 1.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 8.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 4.0 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.2 | 30.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 55.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.2 | 4.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 8.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 2.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 6.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 1.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 2.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 1.6 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 8.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 24.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 18.4 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.1 | 0.9 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 4.4 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 8.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 2.5 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.1 | 1.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.8 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 14.2 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.1 | 1.4 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.1 | 2.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 76.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 4.9 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.4 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.1 | 3.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.2 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 13.7 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 1.9 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 15.7 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 4.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 5.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 3.6 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 1.0 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.4 | 8.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.3 | 14.4 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.2 | 10.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.2 | 32.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.9 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 3.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 2.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 34.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 3.9 | 23.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 2.8 | 8.3 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 2.7 | 5.5 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 2.2 | 13.3 | REACTOME OPSINS | Genes involved in Opsins |
| 1.0 | 15.8 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.8 | 16.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.7 | 14.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.7 | 8.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.4 | 4.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.4 | 7.8 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.4 | 18.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 3.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 5.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 10.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 1.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 9.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.4 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 7.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 2.5 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.1 | 3.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 2.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 2.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |