Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
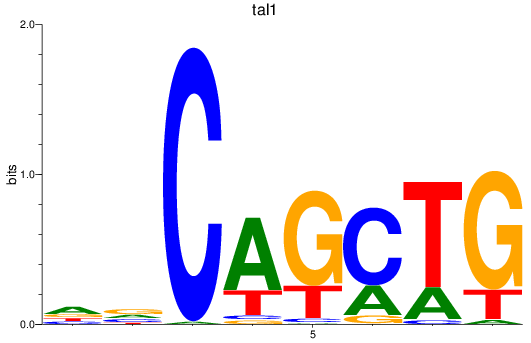
Results for tal1
Z-value: 0.75
Transcription factors associated with tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tal1
|
ENSDARG00000019930 | T-cell acute lymphocytic leukemia 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tal1 | dr11_v1_chr22_+_16535575_16535575 | 0.86 | 1.8e-06 | Click! |
Activity profile of tal1 motif
Sorted Z-values of tal1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_306533 | 1.81 |
ENSDART00000173437
|
poc1a
|
POC1 centriolar protein A |
| chr1_-_59571758 | 1.80 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr21_-_217589 | 1.80 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr21_-_23331619 | 1.67 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr8_-_1051438 | 1.50 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr7_+_44715224 | 1.47 |
ENSDART00000184630
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr22_-_10541372 | 1.45 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr25_-_36261836 | 1.44 |
ENSDART00000179411
|
dus2
|
dihydrouridine synthase 2 |
| chr25_-_36369057 | 1.42 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr15_-_4485828 | 1.41 |
ENSDART00000062868
|
tfdp2
|
transcription factor Dp-2 |
| chr2_+_20866898 | 1.37 |
ENSDART00000150086
|
odr4
|
odr-4 GPCR localization factor homolog |
| chr6_+_40629066 | 1.33 |
ENSDART00000103757
|
slc6a11a
|
solute carrier family 6 (neurotransmitter transporter), member 11a |
| chr14_-_25928899 | 1.29 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr1_+_277731 | 1.26 |
ENSDART00000133431
|
cenpe
|
centromere protein E |
| chr2_+_20793982 | 1.24 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr14_-_22108718 | 1.24 |
ENSDART00000054410
|
med19a
|
mediator complex subunit 19a |
| chr14_-_9281232 | 1.16 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr7_+_5964296 | 1.16 |
ENSDART00000173380
|
si:dkey-23a13.17
|
si:dkey-23a13.17 |
| chr24_-_9300160 | 1.14 |
ENSDART00000152378
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr21_-_31210749 | 1.13 |
ENSDART00000185356
|
zgc:152891
|
zgc:152891 |
| chr3_+_54581987 | 1.12 |
ENSDART00000018071
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr11_+_30296332 | 1.12 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr10_+_16036573 | 1.10 |
ENSDART00000188757
|
lmnb1
|
lamin B1 |
| chr7_+_2455344 | 1.09 |
ENSDART00000172942
|
si:dkey-125e8.4
|
si:dkey-125e8.4 |
| chr7_+_26029672 | 1.07 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr17_-_2590222 | 1.05 |
ENSDART00000185711
|
CR759892.1
|
|
| chr4_+_77943184 | 1.03 |
ENSDART00000159094
|
pacsin2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr3_-_47235997 | 1.02 |
ENSDART00000047071
|
tmed1a
|
transmembrane p24 trafficking protein 1a |
| chr5_+_25762271 | 0.98 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr12_-_48168135 | 0.98 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr23_-_19831739 | 0.98 |
ENSDART00000125066
|
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr17_-_28097760 | 0.97 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr14_-_30390145 | 0.95 |
ENSDART00000045423
|
slc7a2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr17_-_10073926 | 0.94 |
ENSDART00000166081
ENSDART00000161574 |
zgc:109986
|
zgc:109986 |
| chr24_-_6029314 | 0.94 |
ENSDART00000136155
|
ftr60
|
finTRIM family, member 60 |
| chr7_+_6317866 | 0.93 |
ENSDART00000173397
|
si:ch211-220f21.3
|
si:ch211-220f21.3 |
| chr1_-_53468160 | 0.93 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr1_+_292545 | 0.93 |
ENSDART00000148261
|
cenpe
|
centromere protein E |
| chr7_-_6346859 | 0.92 |
ENSDART00000172913
|
si:ch73-368j24.11
|
si:ch73-368j24.11 |
| chr2_+_13694450 | 0.92 |
ENSDART00000077259
ENSDART00000189485 |
ebna1bp2
|
EBNA1 binding protein 2 |
| chr4_+_38990661 | 0.92 |
ENSDART00000133258
|
si:dkey-122c11.1
|
si:dkey-122c11.1 |
| chr17_+_132555 | 0.90 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr8_-_20118549 | 0.89 |
ENSDART00000132218
|
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_+_71683853 | 0.89 |
ENSDART00000163002
|
emilin2b
|
elastin microfibril interfacer 2b |
| chr10_+_8875195 | 0.87 |
ENSDART00000141045
|
itga2.3
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 3 |
| chr18_-_46010 | 0.87 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_-_53880639 | 0.87 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr17_-_2578026 | 0.86 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr9_+_44994214 | 0.86 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr14_+_9485070 | 0.86 |
ENSDART00000161486
ENSDART00000137274 |
st3gal5
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
| chr11_-_22372072 | 0.85 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr3_-_29977495 | 0.84 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr7_+_24049776 | 0.84 |
ENSDART00000166559
|
efs
|
embryonal Fyn-associated substrate |
| chr23_+_20513104 | 0.84 |
ENSDART00000079591
|
dpm1
|
dolichyl-phosphate mannosyltransferase polypeptide 1, catalytic subunit |
| chr4_-_76488581 | 0.83 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr17_+_10074360 | 0.83 |
ENSDART00000166649
|
srp54
|
signal recognition particle 54 |
| chr11_+_6136220 | 0.82 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr22_-_10541712 | 0.82 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr25_+_258883 | 0.82 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr8_+_46327350 | 0.81 |
ENSDART00000145618
|
si:dkey-75a21.2
|
si:dkey-75a21.2 |
| chr2_-_20866758 | 0.80 |
ENSDART00000165374
|
tpra
|
translocated promoter region a, nuclear basket protein |
| chr16_+_54875530 | 0.79 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr1_-_52494122 | 0.78 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr3_+_19665319 | 0.78 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr4_-_2052687 | 0.77 |
ENSDART00000138291
ENSDART00000150844 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr21_+_53504 | 0.77 |
ENSDART00000170452
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr16_+_42772678 | 0.75 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr19_-_24757231 | 0.75 |
ENSDART00000128177
|
si:dkey-154b15.1
|
si:dkey-154b15.1 |
| chr17_-_2595736 | 0.75 |
ENSDART00000128797
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr4_-_67589158 | 0.75 |
ENSDART00000184361
|
BX088712.2
|
|
| chr7_+_56577522 | 0.75 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr5_-_65000312 | 0.75 |
ENSDART00000192893
|
ANXA1 (1 of many)
|
zgc:110283 |
| chr13_+_21674445 | 0.74 |
ENSDART00000100931
ENSDART00000078567 |
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr19_-_1002959 | 0.74 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr25_+_27405738 | 0.74 |
ENSDART00000183266
ENSDART00000115139 |
pot1
|
protection of telomeres 1 homolog |
| chr2_+_49864219 | 0.73 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr8_+_29742237 | 0.73 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr18_-_6862738 | 0.72 |
ENSDART00000192592
|
CT009745.1
|
|
| chr4_-_48636872 | 0.72 |
ENSDART00000168605
|
znf1063
|
zinc finger protein 1063 |
| chr6_-_51573975 | 0.71 |
ENSDART00000073865
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr4_+_64562090 | 0.71 |
ENSDART00000188810
|
si:ch211-223a21.3
|
si:ch211-223a21.3 |
| chr18_+_16717764 | 0.71 |
ENSDART00000166849
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr8_+_39570615 | 0.69 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr1_+_22851261 | 0.68 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr20_+_53474963 | 0.68 |
ENSDART00000138976
|
bub1ba
|
BUB1 mitotic checkpoint serine/threonine kinase Ba |
| chr1_-_59141715 | 0.68 |
ENSDART00000164941
ENSDART00000138870 |
si:ch1073-110a20.1
|
si:ch1073-110a20.1 |
| chr1_-_59232267 | 0.67 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr4_-_52165969 | 0.67 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr17_-_2573021 | 0.67 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr10_-_24753715 | 0.67 |
ENSDART00000192401
|
ilk
|
integrin-linked kinase |
| chr13_+_40770628 | 0.67 |
ENSDART00000085846
|
nkx1.2la
|
NK1 transcription factor related 2-like,a |
| chr23_+_26079467 | 0.67 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr7_-_30127082 | 0.66 |
ENSDART00000173749
|
alpk3b
|
alpha-kinase 3b |
| chr12_-_33789006 | 0.66 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr7_+_33132074 | 0.65 |
ENSDART00000073554
|
zgc:153219
|
zgc:153219 |
| chr8_-_1255321 | 0.65 |
ENSDART00000149605
|
cdc14b
|
cell division cycle 14B |
| chr13_+_1131748 | 0.65 |
ENSDART00000054318
|
wdr92
|
WD repeat domain 92 |
| chr1_+_49686408 | 0.65 |
ENSDART00000140824
|
si:ch211-149l1.2
|
si:ch211-149l1.2 |
| chr19_-_48010490 | 0.64 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr3_+_36671585 | 0.64 |
ENSDART00000159033
|
nde1
|
nudE neurodevelopment protein 1 |
| chr5_-_68782641 | 0.64 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr7_+_42935126 | 0.64 |
ENSDART00000157747
|
BX284696.1
|
|
| chr14_+_9421510 | 0.63 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr6_+_9241121 | 0.63 |
ENSDART00000064989
|
pimr70
|
Pim proto-oncogene, serine/threonine kinase, related 70 |
| chr19_-_27588842 | 0.63 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr21_-_30026359 | 0.63 |
ENSDART00000153645
|
pwwp2a
|
PWWP domain containing 2A |
| chr17_-_2584423 | 0.63 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr4_-_43911462 | 0.62 |
ENSDART00000189056
ENSDART00000162441 ENSDART00000171367 |
znf1104
|
zinc finger protein 1104 |
| chr7_+_56577906 | 0.62 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr1_+_513986 | 0.62 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr7_+_51795667 | 0.62 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr3_-_32590164 | 0.61 |
ENSDART00000151151
|
tspan4b
|
tetraspanin 4b |
| chr22_-_29906764 | 0.60 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr25_-_35142127 | 0.60 |
ENSDART00000049135
|
si:dkey-261m9.12
|
si:dkey-261m9.12 |
| chr7_-_22790630 | 0.59 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr24_+_18714212 | 0.59 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr20_-_51547464 | 0.59 |
ENSDART00000099486
|
disp1
|
dispatched homolog 1 (Drosophila) |
| chr4_+_29206813 | 0.59 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr19_+_43978814 | 0.59 |
ENSDART00000102314
|
nkd3
|
naked cuticle homolog 3 |
| chr19_-_21766461 | 0.58 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr15_-_15227541 | 0.58 |
ENSDART00000184787
|
rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr2_-_59345920 | 0.58 |
ENSDART00000134662
|
ftr37
|
finTRIM family, member 37 |
| chr5_-_68333081 | 0.58 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr12_+_1609563 | 0.58 |
ENSDART00000163559
|
SLC39A11
|
solute carrier family 39 member 11 |
| chr7_+_71535045 | 0.58 |
ENSDART00000047069
|
tyms
|
thymidylate synthetase |
| chr12_+_26632448 | 0.58 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr10_-_1788376 | 0.58 |
ENSDART00000123842
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr11_+_25259058 | 0.57 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr22_-_26353916 | 0.57 |
ENSDART00000077958
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr13_+_35474235 | 0.56 |
ENSDART00000181927
|
mkks
|
McKusick-Kaufman syndrome |
| chr6_+_9893554 | 0.56 |
ENSDART00000064979
|
pimr74
|
Pim proto-oncogene, serine/threonine kinase, related 74 |
| chr1_-_55888970 | 0.56 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr23_+_16807945 | 0.56 |
ENSDART00000080660
|
zgc:114081
|
zgc:114081 |
| chr5_+_58520266 | 0.55 |
ENSDART00000108889
|
oafb
|
OAF homolog b (Drosophila) |
| chr20_-_15062606 | 0.55 |
ENSDART00000175585
ENSDART00000135212 |
prrc2c
|
proline-rich coiled-coil 2C |
| chr20_-_39391833 | 0.55 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr4_-_38033800 | 0.54 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr6_-_42377307 | 0.54 |
ENSDART00000129302
|
emc3
|
ER membrane protein complex subunit 3 |
| chr2_-_59265521 | 0.54 |
ENSDART00000146341
ENSDART00000097799 |
ftr33
|
finTRIM family, member 33 |
| chr19_-_10214264 | 0.54 |
ENSDART00000053300
ENSDART00000148225 |
znf865
|
zinc finger protein 865 |
| chr23_-_36934944 | 0.54 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr4_-_74466703 | 0.53 |
ENSDART00000174032
ENSDART00000189417 |
LO018124.1
|
|
| chr13_-_42530656 | 0.53 |
ENSDART00000084327
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr14_+_28432737 | 0.53 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr23_-_44848961 | 0.53 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr24_+_35827766 | 0.53 |
ENSDART00000144700
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr4_+_77709678 | 0.53 |
ENSDART00000036856
|
AL935186.1
|
|
| chr2_-_5723786 | 0.52 |
ENSDART00000100924
|
cwc25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr7_-_5640822 | 0.52 |
ENSDART00000143767
|
BX000524.1
|
|
| chr16_-_41990421 | 0.51 |
ENSDART00000055921
|
pycard
|
PYD and CARD domain containing |
| chr25_+_1732838 | 0.51 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr15_-_14064302 | 0.51 |
ENSDART00000156384
|
BX927244.1
|
|
| chr23_-_9925568 | 0.51 |
ENSDART00000081268
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr3_-_61377127 | 0.51 |
ENSDART00000155932
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr4_-_20208166 | 0.51 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr18_+_20482369 | 0.50 |
ENSDART00000100668
|
kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4 |
| chr2_-_59157790 | 0.50 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr5_+_41322783 | 0.49 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr4_+_13412030 | 0.49 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr11_-_1392468 | 0.49 |
ENSDART00000004423
|
iars
|
isoleucyl-tRNA synthetase |
| chr5_+_338154 | 0.49 |
ENSDART00000191743
|
rnf170
|
ring finger protein 170 |
| chr20_+_25225112 | 0.49 |
ENSDART00000153088
ENSDART00000127291 ENSDART00000130494 |
moxd1
|
monooxygenase, DBH-like 1 |
| chr15_-_34051457 | 0.49 |
ENSDART00000189764
|
VWDE
|
si:dkey-30e9.7 |
| chr9_-_23747264 | 0.49 |
ENSDART00000141461
ENSDART00000010311 |
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr11_-_5953636 | 0.49 |
ENSDART00000140960
ENSDART00000123601 |
dda1
|
DET1 and DDB1 associated 1 |
| chr15_+_5116179 | 0.48 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr5_+_36895860 | 0.48 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr6_-_29159888 | 0.48 |
ENSDART00000110288
|
zbtb11
|
zinc finger and BTB domain containing 11 |
| chr14_-_36862745 | 0.48 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr9_-_7378566 | 0.48 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr10_-_8294965 | 0.48 |
ENSDART00000167380
|
plpp1a
|
phospholipid phosphatase 1a |
| chr11_-_6970107 | 0.48 |
ENSDART00000171255
|
COMP
|
si:ch211-43f4.1 |
| chr5_+_19933356 | 0.48 |
ENSDART00000088819
|
ankrd13a
|
ankyrin repeat domain 13A |
| chr1_-_29747702 | 0.47 |
ENSDART00000133225
ENSDART00000189670 |
spp2
|
secreted phosphoprotein 2 |
| chr13_-_9024004 | 0.47 |
ENSDART00000169564
|
si:dkey-112g5.11
|
si:dkey-112g5.11 |
| chr23_+_4260458 | 0.47 |
ENSDART00000103747
|
srsf6a
|
serine/arginine-rich splicing factor 6a |
| chr3_+_30500968 | 0.47 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr5_-_43805597 | 0.47 |
ENSDART00000127956
ENSDART00000028099 |
smn1
|
survival of motor neuron 1, telomeric |
| chr16_-_19568795 | 0.47 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr5_+_41477954 | 0.47 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr6_+_27923054 | 0.46 |
ENSDART00000136833
ENSDART00000145533 |
cep63
|
centrosomal protein 63 |
| chr1_+_2260407 | 0.46 |
ENSDART00000058876
|
kpnb3
|
karyopherin (importin) beta 3 |
| chr5_+_65946222 | 0.46 |
ENSDART00000190969
ENSDART00000161578 |
mymk
|
myomaker, myoblast fusion factor |
| chr20_-_3310017 | 0.45 |
ENSDART00000099315
|
cebpz
|
CCAAT/enhancer binding protein (C/EBP), zeta |
| chr2_-_16565690 | 0.45 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr2_-_43739740 | 0.45 |
ENSDART00000113849
|
kif5ba
|
kinesin family member 5B, a |
| chr15_-_47857687 | 0.45 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr23_+_578218 | 0.44 |
ENSDART00000055134
|
ogfr
|
opioid growth factor receptor |
| chr13_+_43639867 | 0.44 |
ENSDART00000042588
ENSDART00000074728 |
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr7_-_17814118 | 0.43 |
ENSDART00000179688
|
ecsit
|
ECSIT signalling integrator |
| chr5_+_30392148 | 0.43 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr25_+_30460378 | 0.43 |
ENSDART00000016310
|
banp
|
BTG3 associated nuclear protein |
| chr9_+_33220342 | 0.43 |
ENSDART00000100893
ENSDART00000113451 |
si:ch211-125e6.13
|
si:ch211-125e6.13 |
| chr19_-_10810006 | 0.43 |
ENSDART00000151157
|
si:dkey-3n22.9
|
si:dkey-3n22.9 |
| chr15_+_23947932 | 0.42 |
ENSDART00000153951
|
myo18ab
|
myosin XVIIIAb |
| chr24_-_26399623 | 0.42 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr3_+_37649436 | 0.42 |
ENSDART00000075039
|
gosr2
|
golgi SNAP receptor complex member 2 |
| chr9_+_33217243 | 0.42 |
ENSDART00000053061
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr20_+_46260580 | 0.42 |
ENSDART00000152872
|
taar14j
|
trace amine associated receptor 14j |
| chr19_+_40122160 | 0.41 |
ENSDART00000143966
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tal1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.3 | 5.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.3 | 0.8 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.3 | 0.8 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.2 | 0.6 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.6 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.2 | 2.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 2.9 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.8 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 0.5 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.5 | GO:0006589 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.2 | 0.9 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 0.5 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.7 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 1.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 1.5 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.7 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 0.7 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 1.7 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 0.7 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.1 | 0.5 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.8 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 1.0 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 0.7 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 0.6 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.1 | 0.7 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.1 | 0.9 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.5 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.6 | GO:0007100 | mitotic centrosome separation(GO:0007100) centrosome separation(GO:0051299) |
| 0.1 | 0.3 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.1 | 0.4 | GO:1904105 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.1 | 1.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.4 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.4 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.1 | 0.4 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.5 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.3 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 1.0 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.3 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 1.0 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 1.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.6 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.9 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.0 | 0.2 | GO:0040016 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) embryonic cleavage(GO:0040016) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 2.9 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.1 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.0 | 0.6 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.5 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.2 | GO:0048569 | post-embryonic organ development(GO:0048569) |
| 0.0 | 0.1 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.7 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.4 | GO:0042177 | negative regulation of protein catabolic process(GO:0042177) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 1.4 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 1.5 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 1.0 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.4 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.8 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.5 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.6 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.8 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.9 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.8 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0032099 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.0 | 0.6 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.2 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0061512 | protein localization to cilium(GO:0061512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.7 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 0.8 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.6 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.0 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.3 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.1 | 0.7 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.6 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 5.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.2 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.6 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.9 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) nuclear periphery(GO:0034399) |
| 0.0 | 1.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.7 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.7 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.4 | GO:0030663 | COPI-coated vesicle membrane(GO:0030663) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.4 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.3 | 0.9 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 2.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 0.8 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.3 | 5.8 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 1.5 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 0.6 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.8 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 0.8 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 0.5 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 1.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 0.9 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.3 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 1.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.7 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.4 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.8 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.3 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.8 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.2 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.9 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.5 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 0.7 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 1.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.7 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 1.5 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 2.0 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.2 | GO:0015923 | mannosidase activity(GO:0015923) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 2.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 2.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 1.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.8 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.0 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |
| 0.0 | 0.6 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.7 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.6 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.3 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.3 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |