Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for stat5a+stat5b
Z-value: 0.71
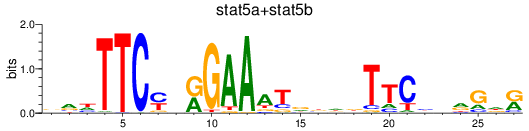
Transcription factors associated with stat5a+stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat5a
|
ENSDARG00000019392 | signal transducer and activator of transcription 5a |
|
stat5b
|
ENSDARG00000055588 | signal transducer and activator of transcription 5b |
|
stat5a
|
ENSDARG00000113871 | signal transducer and activator of transcription 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat5a | dr11_v1_chr3_+_17314997_17314997 | -0.84 | 5.5e-06 | Click! |
| stat5b | dr11_v1_chr12_-_13650344_13650472 | 0.01 | 9.6e-01 | Click! |
Activity profile of stat5a+stat5b motif
Sorted Z-values of stat5a+stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_49819027 | 2.34 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr1_+_230363 | 2.02 |
ENSDART00000153285
|
tfdp1b
|
transcription factor Dp-1, b |
| chr22_-_26005894 | 1.94 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr22_-_26100282 | 1.87 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr18_-_49318823 | 1.70 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr25_+_31323978 | 1.67 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr6_+_45934682 | 1.65 |
ENSDART00000103489
|
cenps
|
centromere protein S |
| chr15_-_23485752 | 1.65 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr3_-_45281350 | 1.60 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr15_+_2857556 | 1.46 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr19_+_15443063 | 1.34 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_-_42035250 | 1.33 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr3_-_32541033 | 1.26 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr18_-_20444296 | 1.18 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr2_-_14793343 | 1.16 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr11_+_30321116 | 1.15 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr20_+_25586099 | 1.15 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr7_+_26029672 | 1.14 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr20_+_28803642 | 1.12 |
ENSDART00000188526
|
fntb
|
farnesyltransferase, CAAX box, beta |
| chr8_+_247163 | 1.06 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr21_-_32284532 | 1.04 |
ENSDART00000190676
|
clk4b
|
CDC-like kinase 4b |
| chr20_+_17581881 | 1.01 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr3_-_46818001 | 0.98 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_-_24521382 | 0.95 |
ENSDART00000092948
|
peli1b
|
pellino E3 ubiquitin protein ligase 1b |
| chr11_+_13024002 | 0.95 |
ENSDART00000104113
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr15_-_44052927 | 0.95 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr24_+_13635108 | 0.94 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr13_+_6092135 | 0.93 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr6_-_37499574 | 0.91 |
ENSDART00000023163
|
jade3
|
jade family PHD finger 3 |
| chr16_+_25171832 | 0.90 |
ENSDART00000156416
|
wu:fe05a04
|
wu:fe05a04 |
| chr16_-_41787421 | 0.90 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr16_+_9762261 | 0.90 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr7_+_17106160 | 0.88 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr21_+_37090585 | 0.84 |
ENSDART00000182971
|
znf346
|
zinc finger protein 346 |
| chr6_+_16870004 | 0.84 |
ENSDART00000154794
|
pimr21
|
Pim proto-oncogene, serine/threonine kinase, related 21 |
| chr5_+_32141790 | 0.82 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr4_-_49952636 | 0.81 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr10_-_8434816 | 0.81 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr22_-_9861531 | 0.80 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr7_-_38687431 | 0.79 |
ENSDART00000074490
|
aplnr2
|
apelin receptor 2 |
| chr14_+_6953244 | 0.78 |
ENSDART00000159470
|
rack1
|
receptor for activated C kinase 1 |
| chr17_-_43517542 | 0.78 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr2_-_42109575 | 0.77 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr17_-_6536305 | 0.76 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr1_+_33322555 | 0.75 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr9_-_49860756 | 0.73 |
ENSDART00000044270
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr4_-_1818315 | 0.72 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr6_-_43283122 | 0.72 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr5_+_29160324 | 0.70 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr11_+_16040517 | 0.70 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr15_-_25083200 | 0.66 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr19_+_7759354 | 0.65 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr17_-_6536466 | 0.63 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr15_+_37412883 | 0.58 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr8_-_28342487 | 0.56 |
ENSDART00000108508
|
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr6_+_16895685 | 0.56 |
ENSDART00000154090
|
pimr28
|
Pim proto-oncogene, serine/threonine kinase, related 28 |
| chr8_-_31053872 | 0.56 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr21_-_14692119 | 0.55 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr8_+_25959940 | 0.54 |
ENSDART00000143011
ENSDART00000140626 |
si:dkey-72l14.4
|
si:dkey-72l14.4 |
| chr20_+_48754045 | 0.54 |
ENSDART00000062578
|
zgc:110348
|
zgc:110348 |
| chr14_+_22132388 | 0.53 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr4_+_13586455 | 0.52 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr15_+_29088426 | 0.51 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr8_-_47212686 | 0.51 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr17_+_46864116 | 0.51 |
ENSDART00000156250
|
pimr27
|
Pim proto-oncogene, serine/threonine kinase, related 27 |
| chr19_-_43757568 | 0.48 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr24_-_25166720 | 0.47 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr17_+_12255642 | 0.47 |
ENSDART00000155175
|
pimr175
|
Pim proto-oncogene, serine/threonine kinase, related 175 |
| chr8_-_36618073 | 0.46 |
ENSDART00000047912
|
gpkow
|
G patch domain and KOW motifs |
| chr20_+_37294112 | 0.45 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr3_-_46817838 | 0.45 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr3_-_6608342 | 0.44 |
ENSDART00000161188
|
si:ch73-59f11.3
|
si:ch73-59f11.3 |
| chr14_-_6987649 | 0.43 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr6_-_3998199 | 0.42 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr2_-_22659450 | 0.42 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr20_-_3238110 | 0.41 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr17_+_11372531 | 0.41 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr4_-_1497384 | 0.41 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr6_-_53048291 | 0.40 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr11_+_16040680 | 0.39 |
ENSDART00000157703
|
agtrap
|
angiotensin II receptor-associated protein |
| chr6_-_18393476 | 0.38 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr15_-_3282220 | 0.38 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr15_-_32383529 | 0.35 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr5_-_42878178 | 0.35 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr1_+_57040472 | 0.35 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr7_+_17560808 | 0.33 |
ENSDART00000159699
ENSDART00000165724 ENSDART00000129017 ENSDART00000101955 |
nitr1b
|
novel immune-type receptor 1b |
| chr25_+_34749812 | 0.32 |
ENSDART00000185712
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr9_+_18572970 | 0.31 |
ENSDART00000114860
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr7_+_17303944 | 0.29 |
ENSDART00000163505
ENSDART00000162169 ENSDART00000161291 |
nitr9
|
novel immune-type receptor 9 |
| chr10_-_7821686 | 0.28 |
ENSDART00000121531
|
mat2aa
|
methionine adenosyltransferase II, alpha a |
| chr4_+_57580303 | 0.28 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr5_-_16791223 | 0.27 |
ENSDART00000192744
|
atad1a
|
ATPase family, AAA domain containing 1a |
| chr8_+_19548985 | 0.26 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr6_-_37749711 | 0.26 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr15_-_31401361 | 0.25 |
ENSDART00000124360
|
or111-7
|
odorant receptor, family D, subfamily 111, member 7 |
| chr15_-_2857961 | 0.25 |
ENSDART00000033263
|
ankrd49
|
ankyrin repeat domain 49 |
| chr15_-_25010400 | 0.24 |
ENSDART00000155236
|
si:ch211-198e20.10
|
si:ch211-198e20.10 |
| chr1_+_33322174 | 0.23 |
ENSDART00000140435
|
mxra5a
|
matrix-remodelling associated 5a |
| chr15_-_15449929 | 0.22 |
ENSDART00000101918
|
proca1
|
protein interacting with cyclin A1 |
| chr15_-_29556757 | 0.22 |
ENSDART00000060049
|
hspa13
|
heat shock protein 70 family, member 13 |
| chr4_+_34599803 | 0.20 |
ENSDART00000134088
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr24_-_25166416 | 0.19 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr16_+_25126935 | 0.19 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr22_-_16180467 | 0.18 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr17_-_46933567 | 0.18 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr8_+_10920205 | 0.18 |
ENSDART00000179742
|
BX321886.1
|
|
| chr2_-_37140423 | 0.18 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr10_+_36299541 | 0.18 |
ENSDART00000166138
|
or106-7
|
odorant receptor, family G, subfamily 106, member 7 |
| chr7_+_34786591 | 0.18 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr17_-_46817295 | 0.17 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr13_+_2861265 | 0.17 |
ENSDART00000170602
ENSDART00000171687 |
XPO5
|
si:ch211-233m11.2 |
| chr4_-_57571688 | 0.17 |
ENSDART00000160179
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr15_-_25039083 | 0.16 |
ENSDART00000154409
|
pimr190
|
Pim proto-oncogene, serine/threonine kinase, related 190 |
| chr6_-_17266221 | 0.16 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr17_-_24439672 | 0.16 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr24_+_31374324 | 0.15 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr5_+_20366453 | 0.15 |
ENSDART00000193141
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr24_+_25032340 | 0.15 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr2_+_37140448 | 0.15 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr10_+_44700103 | 0.14 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr17_+_44463230 | 0.14 |
ENSDART00000130311
|
naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr5_-_4245186 | 0.14 |
ENSDART00000145912
|
si:ch211-283g2.3
|
si:ch211-283g2.3 |
| chr15_-_6966221 | 0.13 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr20_+_28803977 | 0.13 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr4_-_12323228 | 0.11 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr7_+_58751504 | 0.11 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr6_-_17052116 | 0.11 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr4_+_47174366 | 0.10 |
ENSDART00000170732
|
si:dkey-26m3.1
|
si:dkey-26m3.1 |
| chr22_+_36914636 | 0.09 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr4_+_49322310 | 0.09 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr17_+_6536152 | 0.08 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr23_+_26017227 | 0.08 |
ENSDART00000002939
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr6_-_16735402 | 0.07 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr10_-_43568239 | 0.07 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr4_-_57572054 | 0.07 |
ENSDART00000158610
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr11_-_17964525 | 0.07 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr8_-_46060685 | 0.06 |
ENSDART00000121902
|
pimr188
|
Pim proto-oncogene, serine/threonine kinase, related 188 |
| chr6_+_39923052 | 0.06 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr7_+_29890292 | 0.04 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr20_-_54508650 | 0.03 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr7_-_13362590 | 0.03 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr6_-_16804001 | 0.03 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr7_+_53254234 | 0.03 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr15_-_32383340 | 0.01 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr3_+_35608385 | 0.00 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat5a+stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.8 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.3 | 1.0 | GO:0090248 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.3 | 1.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 1.1 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.3 | 0.8 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.7 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.2 | 1.0 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 1.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.4 | GO:0039535 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.4 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.7 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.4 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 1.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.9 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.1 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.6 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.8 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 2.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.7 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.9 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.5 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 1.1 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0051182 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.2 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.1 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.0 | 4.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.6 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.3 | 1.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 1.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.7 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.7 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 1.0 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.2 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 0.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.2 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.8 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 1.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.8 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.6 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 1.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.3 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 2.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.1 | 0.9 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.7 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.4 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.1 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.7 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.8 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 1.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 1.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 2.0 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |