Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
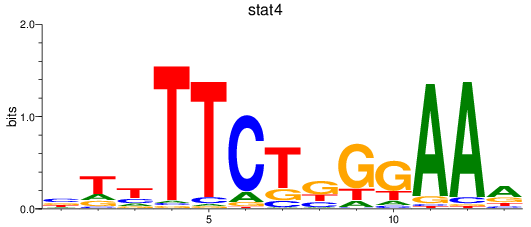
Results for stat4
Z-value: 0.47
Transcription factors associated with stat4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat4
|
ENSDARG00000028731 | signal transducer and activator of transcription 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat4 | dr11_v1_chr9_+_41156287_41156287 | -0.24 | 3.1e-01 | Click! |
Activity profile of stat4 motif
Sorted Z-values of stat4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_10098191 | 0.79 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr7_+_48870496 | 0.59 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr21_+_20901505 | 0.56 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr16_+_50289916 | 0.53 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr6_+_21005725 | 0.53 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr8_+_39795918 | 0.52 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr1_-_38171648 | 0.50 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr2_+_42005217 | 0.48 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr21_-_40049642 | 0.45 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr2_+_26288301 | 0.44 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr8_-_18203092 | 0.44 |
ENSDART00000140620
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr4_+_4803698 | 0.44 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr2_-_9989919 | 0.44 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr2_+_4402765 | 0.43 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr6_-_7439490 | 0.42 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr16_-_21785261 | 0.42 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr4_+_76735113 | 0.41 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr18_+_30567945 | 0.40 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr2_-_42035250 | 0.38 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr25_+_20081553 | 0.38 |
ENSDART00000174684
|
tnni4b.1
|
troponin I4b, tandem duplicate 1 |
| chr7_+_45963767 | 0.37 |
ENSDART00000163098
|
pop4
|
POP4 homolog, ribonuclease P/MRP subunit |
| chr19_-_2317558 | 0.37 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr2_+_29257942 | 0.37 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr21_-_32060993 | 0.36 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr5_+_37978501 | 0.35 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr2_+_2957515 | 0.34 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr22_-_13350240 | 0.34 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr7_-_26518086 | 0.34 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr21_-_25565392 | 0.33 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr11_+_19603251 | 0.32 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr7_-_24875421 | 0.32 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr6_-_8377055 | 0.32 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr8_+_8937723 | 0.31 |
ENSDART00000145970
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr8_-_25033681 | 0.31 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr23_+_43718115 | 0.30 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr17_+_30591287 | 0.30 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr2_+_16449627 | 0.30 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr23_-_31763753 | 0.30 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr4_+_18960914 | 0.30 |
ENSDART00000139730
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr21_-_11970199 | 0.29 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr23_-_43718067 | 0.29 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr4_+_76637548 | 0.29 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr23_-_38160024 | 0.29 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr23_+_17417539 | 0.28 |
ENSDART00000182605
|
BX649300.2
|
|
| chr3_-_13599482 | 0.28 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr21_+_19834072 | 0.28 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr15_+_20529197 | 0.28 |
ENSDART00000060935
ENSDART00000137926 ENSDART00000140087 |
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr2_+_4207209 | 0.27 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr17_-_10838434 | 0.27 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr7_+_29065915 | 0.27 |
ENSDART00000136657
|
vrk3
|
vaccinia related kinase 3 |
| chr16_-_31718013 | 0.27 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr13_-_31647323 | 0.27 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr12_+_28888975 | 0.27 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr6_-_30689126 | 0.26 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr16_+_27564270 | 0.26 |
ENSDART00000140460
|
tmem67
|
transmembrane protein 67 |
| chr15_+_33691455 | 0.26 |
ENSDART00000170329
|
hdac12
|
histone deacetylase 12 |
| chr19_+_28284120 | 0.26 |
ENSDART00000103862
|
mrpl36
|
mitochondrial ribosomal protein L36 |
| chr1_-_36770883 | 0.26 |
ENSDART00000167831
|
prmt9
|
protein arginine methyltransferase 9 |
| chr3_-_45281350 | 0.26 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr9_+_15890558 | 0.26 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr1_-_18585046 | 0.26 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr15_+_25489406 | 0.26 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr20_+_35433192 | 0.25 |
ENSDART00000017485
|
sf3b6
|
splicing factor 3b, subunit 6 |
| chr22_+_24446964 | 0.25 |
ENSDART00000166812
|
p3h2
|
prolyl 3-hydroxylase 2 |
| chr9_-_27738110 | 0.24 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr25_+_16312258 | 0.24 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr8_-_18203274 | 0.24 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr16_-_9830451 | 0.24 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr6_+_29386728 | 0.24 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr11_+_36777613 | 0.24 |
ENSDART00000065644
ENSDART00000158151 |
parapinopsinb
|
parapinopsin b |
| chr17_+_32571584 | 0.24 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr5_-_22027357 | 0.24 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr17_-_43517542 | 0.23 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr18_+_7543347 | 0.23 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr7_-_27696958 | 0.23 |
ENSDART00000173470
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr22_+_26263290 | 0.23 |
ENSDART00000184840
|
BX649315.1
|
|
| chr7_+_65673885 | 0.23 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr10_+_8401929 | 0.22 |
ENSDART00000059028
|
hvcn1
|
hydrogen voltage-gated channel 1 |
| chr16_-_27564256 | 0.22 |
ENSDART00000078297
|
zgc:153215
|
zgc:153215 |
| chr19_-_3821678 | 0.22 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr8_+_44613135 | 0.22 |
ENSDART00000063392
|
lsm1
|
LSM1, U6 small nuclear RNA associated |
| chr19_+_10331325 | 0.22 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr5_+_36895860 | 0.21 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr5_-_37959874 | 0.21 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr16_-_31717851 | 0.21 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr21_+_6397463 | 0.21 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr5_+_23256187 | 0.21 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr19_-_2318391 | 0.21 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr7_-_29043075 | 0.21 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr19_-_22843480 | 0.21 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr5_-_57624425 | 0.20 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr2_+_42005475 | 0.20 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr9_+_44994214 | 0.20 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr7_-_28696556 | 0.20 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr23_+_4350897 | 0.20 |
ENSDART00000190593
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr23_-_5783421 | 0.20 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr17_-_45386546 | 0.20 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr14_-_33278084 | 0.20 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr3_-_52661242 | 0.20 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr11_+_43401592 | 0.20 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr3_-_43954971 | 0.20 |
ENSDART00000167159
ENSDART00000189315 |
ubfd1
|
ubiquitin family domain containing 1 |
| chr22_+_20172018 | 0.20 |
ENSDART00000188104
|
hmg20b
|
high mobility group 20B |
| chr21_+_8341774 | 0.20 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr10_-_34033680 | 0.19 |
ENSDART00000180734
|
rfc3
|
replication factor C (activator 1) 3 |
| chr11_+_43401201 | 0.19 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr7_+_32693890 | 0.19 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr24_-_17039638 | 0.19 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr7_-_26497947 | 0.19 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr3_+_40289418 | 0.19 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr20_+_30578163 | 0.19 |
ENSDART00000159485
|
fgfr1op
|
FGFR1 oncogene partner |
| chr2_-_15031858 | 0.18 |
ENSDART00000191478
|
hccsa.1
|
holocytochrome c synthase a |
| chr11_+_8152872 | 0.18 |
ENSDART00000091638
ENSDART00000138057 ENSDART00000166379 |
samd13
|
sterile alpha motif domain containing 13 |
| chr5_+_54685175 | 0.18 |
ENSDART00000115016
|
pmchl
|
pro-melanin-concentrating hormone, like |
| chr13_+_8255106 | 0.18 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr4_-_61920018 | 0.18 |
ENSDART00000164832
|
znf1056
|
zinc finger protein 1056 |
| chr15_-_31390985 | 0.18 |
ENSDART00000060117
|
or111-6
|
odorant receptor, family D, subfamily 111, member 6 |
| chr6_-_3998199 | 0.18 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr19_-_27334394 | 0.18 |
ENSDART00000052359
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr24_+_18299506 | 0.18 |
ENSDART00000172056
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr22_-_11078988 | 0.18 |
ENSDART00000126664
ENSDART00000006927 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr22_-_9861531 | 0.17 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr16_+_38394371 | 0.17 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr1_+_12195700 | 0.17 |
ENSDART00000040307
|
tdrd7a
|
tudor domain containing 7 a |
| chr13_+_2908764 | 0.17 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr24_+_13635108 | 0.17 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr15_-_25083200 | 0.17 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr20_+_23960525 | 0.17 |
ENSDART00000042123
|
cx52.6
|
connexin 52.6 |
| chr7_+_34487833 | 0.17 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr2_-_27330181 | 0.17 |
ENSDART00000137053
|
tmx3a
|
thioredoxin related transmembrane protein 3a |
| chr4_+_4232562 | 0.17 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr6_+_18423402 | 0.17 |
ENSDART00000159747
|
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr12_-_17602958 | 0.17 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr17_-_45387134 | 0.17 |
ENSDART00000010975
|
tmem206
|
transmembrane protein 206 |
| chr12_-_9796237 | 0.17 |
ENSDART00000149344
|
prdm9
|
PR domain containing 9 |
| chr15_-_31390760 | 0.17 |
ENSDART00000190774
|
or111-6
|
odorant receptor, family D, subfamily 111, member 6 |
| chr18_-_29977431 | 0.17 |
ENSDART00000135357
|
si:ch211-220f16.2
|
si:ch211-220f16.2 |
| chr19_+_7938121 | 0.17 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr15_-_26538989 | 0.16 |
ENSDART00000032880
|
rpa1
|
replication protein A1 |
| chr15_-_18176694 | 0.16 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr4_+_14971239 | 0.16 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr25_+_19489370 | 0.16 |
ENSDART00000091132
|
glsl
|
glutaminase like |
| chr14_+_28473173 | 0.16 |
ENSDART00000017075
|
xiap
|
X-linked inhibitor of apoptosis |
| chr20_-_28433616 | 0.15 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr21_+_25777425 | 0.15 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr5_+_25072894 | 0.15 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr3_+_24275766 | 0.15 |
ENSDART00000055607
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr15_+_11883804 | 0.15 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr16_-_10071886 | 0.15 |
ENSDART00000136168
ENSDART00000130382 |
ptpn2a
|
protein tyrosine phosphatase, non-receptor type 2, a |
| chr24_-_37568359 | 0.15 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr20_+_46513651 | 0.15 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr6_-_22369125 | 0.15 |
ENSDART00000083038
|
nprl2
|
NPR2 like, GATOR1 complex subunit |
| chr24_-_26485098 | 0.15 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr7_-_25049377 | 0.15 |
ENSDART00000077191
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr13_+_13681681 | 0.15 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr23_-_33558161 | 0.14 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr5_+_54280135 | 0.14 |
ENSDART00000170218
ENSDART00000166050 |
rangrf
|
RAN guanine nucleotide release factor |
| chr7_+_12950507 | 0.14 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr6_+_30689239 | 0.14 |
ENSDART00000065206
|
wdr78
|
WD repeat domain 78 |
| chr5_-_42878178 | 0.14 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr9_+_31282161 | 0.14 |
ENSDART00000010774
|
zic2a
|
zic family member 2 (odd-paired homolog, Drosophila), a |
| chr21_+_39197418 | 0.14 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr18_-_20444296 | 0.14 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr3_-_3439150 | 0.13 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr17_-_45386823 | 0.13 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr5_+_36895545 | 0.13 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr21_+_20488662 | 0.13 |
ENSDART00000147951
|
efna5a
|
ephrin-A5a |
| chr25_-_27842654 | 0.13 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr8_-_47212686 | 0.13 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr22_-_28645676 | 0.13 |
ENSDART00000005883
|
jagn1b
|
jagunal homolog 1b |
| chr7_+_33133547 | 0.13 |
ENSDART00000191967
|
si:ch211-194p6.7
|
si:ch211-194p6.7 |
| chr6_-_11800427 | 0.12 |
ENSDART00000126243
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr2_+_37838259 | 0.12 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr14_-_22015232 | 0.12 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr4_-_1818315 | 0.12 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr4_-_12725513 | 0.12 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr13_-_49819027 | 0.12 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr1_-_28831848 | 0.12 |
ENSDART00000148536
|
GK3P
|
zgc:172295 |
| chr25_+_32390794 | 0.12 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr6_-_54179860 | 0.11 |
ENSDART00000164283
|
rps10
|
ribosomal protein S10 |
| chr5_+_43870389 | 0.11 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr7_+_4854690 | 0.11 |
ENSDART00000131645
|
si:dkey-28d5.7
|
si:dkey-28d5.7 |
| chr6_-_22064158 | 0.11 |
ENSDART00000153585
|
cfap100
|
cilia and flagella associated protein 100 |
| chr16_-_11859309 | 0.11 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr11_+_41459408 | 0.11 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr9_-_9225980 | 0.11 |
ENSDART00000180301
|
cbsb
|
cystathionine-beta-synthase b |
| chr15_-_5239536 | 0.11 |
ENSDART00000081704
|
or128-2
|
odorant receptor, family E, subfamily 128, member 2 |
| chr19_+_43885770 | 0.11 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr15_-_5230423 | 0.11 |
ENSDART00000174322
|
or128-3
|
odorant receptor, family E, subfamily 128, member 3 |
| chr14_-_14566417 | 0.10 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr14_+_34780886 | 0.10 |
ENSDART00000130469
ENSDART00000188806 ENSDART00000172924 ENSDART00000173266 ENSDART00000166377 ENSDART00000173371 |
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr15_-_23783822 | 0.10 |
ENSDART00000180932
|
rad1
|
RAD1 homolog (S. pombe) |
| chr8_+_41413919 | 0.10 |
ENSDART00000142590
|
si:ch211-152p23.2
|
si:ch211-152p23.2 |
| chr21_+_39197628 | 0.10 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr3_+_3852614 | 0.10 |
ENSDART00000184334
ENSDART00000162313 |
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr15_-_18200358 | 0.10 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr5_+_67835595 | 0.10 |
ENSDART00000159386
|
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr12_+_15078124 | 0.10 |
ENSDART00000187571
|
CR456635.1
|
|
| chr8_-_48704688 | 0.10 |
ENSDART00000165307
|
pimr182
|
Pim proto-oncogene, serine/threonine kinase, related 182 |
| chr22_+_20427170 | 0.09 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr18_+_17725410 | 0.09 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.4 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.1 | 0.3 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.1 | 0.4 | GO:0060406 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.5 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.4 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.2 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 0.2 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.2 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.1 | 0.2 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 0.2 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.2 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.3 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.1 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.2 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.2 | GO:0017006 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.1 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.0 | 0.6 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.2 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.0 | 0.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0002836 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.3 | GO:0072531 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine-containing compound transmembrane transport(GO:0072531) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.4 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.0 | GO:1904478 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.5 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.3 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.0 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.0 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.2 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.2 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.1 | 0.2 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.1 | 0.3 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.3 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |