Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
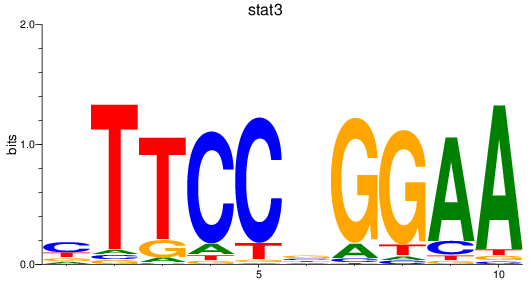
Results for stat3
Z-value: 0.56
Transcription factors associated with stat3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat3
|
ENSDARG00000022712 | signal transducer and activator of transcription 3 (acute-phase response factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat3 | dr11_v1_chr3_+_17030665_17030704 | -0.76 | 1.8e-04 | Click! |
Activity profile of stat3 motif
Sorted Z-values of stat3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16595177 | 2.68 |
ENSDART00000133962
|
si:dkey-239j18.2
|
si:dkey-239j18.2 |
| chr12_-_16941319 | 2.58 |
ENSDART00000109968
|
zgc:174855
|
zgc:174855 |
| chr12_+_16949391 | 2.26 |
ENSDART00000152635
|
zgc:174153
|
zgc:174153 |
| chr19_+_10846182 | 1.53 |
ENSDART00000168479
|
apoa4a
|
apolipoprotein A-IV a |
| chr13_+_33651416 | 1.45 |
ENSDART00000180221
|
BX005372.1
|
|
| chr13_+_14976108 | 1.41 |
ENSDART00000011520
|
noto
|
notochord homeobox |
| chr19_-_47782916 | 1.35 |
ENSDART00000063337
|
cdca8
|
cell division cycle associated 8 |
| chr23_+_42414652 | 1.34 |
ENSDART00000171119
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr19_-_47782586 | 1.27 |
ENSDART00000177126
|
cdca8
|
cell division cycle associated 8 |
| chr21_-_34844059 | 1.14 |
ENSDART00000136402
|
zgc:56585
|
zgc:56585 |
| chr21_-_34844316 | 1.12 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr19_+_15443540 | 1.09 |
ENSDART00000193355
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_13710439 | 1.04 |
ENSDART00000155712
|
ebna1bp2
|
EBNA1 binding protein 2 |
| chr21_+_20901505 | 1.04 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr14_-_32744464 | 1.00 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr24_+_31361407 | 0.99 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr15_+_24737599 | 0.97 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr13_-_49819027 | 0.96 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr21_+_8341774 | 0.94 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr5_-_23619911 | 0.92 |
ENSDART00000020004
|
mtmr2
|
myotubularin related protein 2 |
| chr19_+_15443063 | 0.92 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_15443353 | 0.90 |
ENSDART00000135923
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr19_+_10845953 | 0.89 |
ENSDART00000157589
|
apoa4a
|
apolipoprotein A-IV a |
| chr7_+_41313568 | 0.89 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr21_+_43328685 | 0.85 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr7_+_38809241 | 0.84 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr11_+_12720171 | 0.81 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_+_26288301 | 0.80 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr18_-_26797723 | 0.79 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr25_-_24240797 | 0.76 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr4_+_47257854 | 0.74 |
ENSDART00000173868
|
crestin
|
crestin |
| chr5_+_24179307 | 0.73 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr17_-_10838434 | 0.71 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr13_+_39532050 | 0.69 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr10_-_20105553 | 0.69 |
ENSDART00000184502
|
pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr17_+_16046314 | 0.67 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr19_+_10937932 | 0.67 |
ENSDART00000168968
|
si:ch73-347e22.8
|
si:ch73-347e22.8 |
| chr7_+_22313533 | 0.67 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr18_-_14860435 | 0.66 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr9_-_43644261 | 0.66 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr7_-_22790630 | 0.65 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr10_+_13209580 | 0.65 |
ENSDART00000000887
ENSDART00000136932 |
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr2_-_37837472 | 0.64 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr14_+_11457500 | 0.64 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr12_-_34435604 | 0.64 |
ENSDART00000115088
|
birc5a
|
baculoviral IAP repeat containing 5a |
| chr15_-_3282220 | 0.63 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr4_-_1818315 | 0.62 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr17_+_10318071 | 0.62 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr3_-_35541378 | 0.62 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr10_-_3332362 | 0.59 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr2_-_37896646 | 0.59 |
ENSDART00000075931
|
hbl1
|
hexose-binding lectin 1 |
| chr5_+_28271412 | 0.59 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr3_+_19665319 | 0.59 |
ENSDART00000007857
ENSDART00000193509 |
mettl2a
|
methyltransferase like 2A |
| chr7_-_28696556 | 0.59 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr6_+_55357188 | 0.59 |
ENSDART00000158219
|
pltp
|
phospholipid transfer protein |
| chr15_-_1485086 | 0.59 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr23_-_5783421 | 0.57 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr13_-_12021566 | 0.57 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr18_-_19004247 | 0.57 |
ENSDART00000135800
|
ints14
|
integrator complex subunit 14 |
| chr17_+_16046132 | 0.57 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr22_-_834106 | 0.57 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr19_+_43359075 | 0.56 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr6_+_43015916 | 0.56 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr1_-_26398361 | 0.56 |
ENSDART00000160183
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr16_-_31718013 | 0.55 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr19_+_47783137 | 0.55 |
ENSDART00000024777
ENSDART00000158979 |
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr5_+_25765522 | 0.55 |
ENSDART00000133217
|
tmem2
|
transmembrane protein 2 |
| chr15_-_23475051 | 0.55 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr5_+_65040228 | 0.55 |
ENSDART00000164278
|
pmpca
|
peptidase (mitochondrial processing) alpha |
| chr3_+_40289418 | 0.54 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr16_-_22930925 | 0.54 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr15_-_1484795 | 0.54 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr2_+_37837249 | 0.53 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr5_+_15819651 | 0.53 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr2_-_47431205 | 0.52 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr16_+_42772678 | 0.52 |
ENSDART00000155575
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr15_+_40188076 | 0.52 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr23_-_33558161 | 0.52 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr6_-_7726849 | 0.52 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr9_+_40825065 | 0.52 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr1_-_59313465 | 0.51 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr21_+_19834072 | 0.49 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr8_+_32755688 | 0.49 |
ENSDART00000137897
|
hmcn2
|
hemicentin 2 |
| chr18_-_3166726 | 0.49 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr2_+_4402765 | 0.48 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr5_+_36895860 | 0.48 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr7_-_22941472 | 0.47 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr2_-_37837056 | 0.47 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr2_+_10766744 | 0.46 |
ENSDART00000015379
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr20_+_35433192 | 0.46 |
ENSDART00000017485
|
sf3b6
|
splicing factor 3b, subunit 6 |
| chr18_+_37219140 | 0.46 |
ENSDART00000088247
|
qpctla
|
glutaminyl-peptide cyclotransferase-like a |
| chr23_-_10175898 | 0.46 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr23_+_5524247 | 0.46 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr20_-_23440955 | 0.46 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr6_+_36821621 | 0.45 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr1_-_17711636 | 0.45 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr20_+_27087539 | 0.45 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr2_+_20967673 | 0.45 |
ENSDART00000057174
|
arpc5a
|
actin related protein 2/3 complex, subunit 5A |
| chr3_+_18050667 | 0.44 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr16_-_31717851 | 0.44 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr13_+_22719789 | 0.43 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr8_-_14609284 | 0.43 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr6_-_34220641 | 0.42 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr2_+_44972720 | 0.42 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr8_+_29749017 | 0.41 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr16_+_48631412 | 0.41 |
ENSDART00000154273
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr2_+_17181777 | 0.41 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr21_+_35215810 | 0.41 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr24_+_16149251 | 0.41 |
ENSDART00000188289
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr16_+_31922065 | 0.40 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr16_+_31921812 | 0.40 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr13_+_25549425 | 0.40 |
ENSDART00000087553
ENSDART00000169199 |
sec23ip
|
SEC23 interacting protein |
| chr24_+_37406535 | 0.40 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr22_-_9861531 | 0.39 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr24_+_16149514 | 0.38 |
ENSDART00000152087
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr23_+_30736895 | 0.38 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr23_-_19715799 | 0.38 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr9_-_12659140 | 0.38 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr14_-_6987649 | 0.37 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr20_-_5400395 | 0.37 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr13_-_25548733 | 0.37 |
ENSDART00000168099
ENSDART00000135788 ENSDART00000077655 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr6_-_35106425 | 0.37 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr2_+_41526904 | 0.36 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr16_-_32006737 | 0.36 |
ENSDART00000184813
ENSDART00000179827 |
gstk4
|
glutathione S-transferase kappa 4 |
| chr18_+_18982077 | 0.36 |
ENSDART00000006300
|
hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr11_-_287670 | 0.36 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr3_-_6709938 | 0.35 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr6_-_40573221 | 0.35 |
ENSDART00000157203
ENSDART00000154951 |
tomm6
|
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr6_+_13232934 | 0.34 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr11_+_16040517 | 0.33 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr9_+_20853894 | 0.33 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr1_+_32341734 | 0.32 |
ENSDART00000135078
|
pudp
|
pseudouridine 5'-phosphatase |
| chr4_-_72356954 | 0.32 |
ENSDART00000127023
|
si:cabz01071909.2
|
si:cabz01071909.2 |
| chr14_+_11395383 | 0.32 |
ENSDART00000106657
|
glod5
|
glyoxalase domain containing 5 |
| chr25_-_28587896 | 0.32 |
ENSDART00000111908
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr14_+_7932973 | 0.32 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr3_+_59899452 | 0.31 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr13_+_6092135 | 0.31 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr24_+_13635108 | 0.31 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr19_-_30420602 | 0.31 |
ENSDART00000144121
|
ppp1r10
|
protein phosphatase 1, regulatory subunit 10 |
| chr11_-_20956309 | 0.30 |
ENSDART00000188659
|
CABZ01008739.1
|
|
| chr8_-_30979494 | 0.30 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr1_+_29741843 | 0.30 |
ENSDART00000136066
|
raph1b
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1b |
| chr5_+_29820266 | 0.30 |
ENSDART00000146331
ENSDART00000098315 |
f11r.2
|
F11 receptor, tandem duplicate 2 |
| chr6_+_40573365 | 0.30 |
ENSDART00000004075
|
uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr20_+_32497260 | 0.30 |
ENSDART00000145175
ENSDART00000132921 |
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr21_+_39673157 | 0.30 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr3_+_1219344 | 0.30 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr5_+_67835595 | 0.30 |
ENSDART00000159386
|
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr18_-_15373620 | 0.30 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr1_+_51827046 | 0.30 |
ENSDART00000052992
|
dand5
|
DAN domain family, member 5 |
| chr22_+_10713713 | 0.29 |
ENSDART00000122349
|
hiat1b
|
hippocampus abundant transcript 1b |
| chr4_+_3455665 | 0.29 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr9_-_20853439 | 0.29 |
ENSDART00000028247
ENSDART00000133321 |
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr17_+_34206167 | 0.29 |
ENSDART00000136167
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr3_+_4502066 | 0.29 |
ENSDART00000088610
|
rangap1a
|
RAN GTPase activating protein 1a |
| chr18_+_44631789 | 0.29 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr16_-_15387459 | 0.29 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr19_+_5418006 | 0.28 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr21_-_5879897 | 0.28 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr14_-_21661015 | 0.28 |
ENSDART00000189403
ENSDART00000172442 ENSDART00000181913 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr9_+_26103814 | 0.28 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr11_-_17964525 | 0.28 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr6_+_37655078 | 0.28 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr5_+_26213874 | 0.27 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr14_-_24101897 | 0.27 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr22_+_9239831 | 0.26 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr18_+_6054816 | 0.26 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr21_-_43457554 | 0.26 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr1_+_5402476 | 0.26 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr6_+_33897624 | 0.25 |
ENSDART00000166706
|
ccdc17
|
coiled-coil domain containing 17 |
| chr4_+_29206813 | 0.25 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr8_-_26033176 | 0.25 |
ENSDART00000184533
|
BX784025.1
|
|
| chr20_-_3997531 | 0.25 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr20_-_20932760 | 0.25 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr4_+_76306625 | 0.25 |
ENSDART00000193823
ENSDART00000180622 |
si:ch73-389k6.1
|
si:ch73-389k6.1 |
| chr7_-_29043075 | 0.25 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr4_-_4592287 | 0.25 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr22_+_11153590 | 0.24 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr1_-_11616295 | 0.24 |
ENSDART00000188891
|
si:dkey-26i13.5
|
si:dkey-26i13.5 |
| chr18_+_22217489 | 0.23 |
ENSDART00000165464
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr16_-_19254323 | 0.23 |
ENSDART00000138744
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr16_+_25343934 | 0.23 |
ENSDART00000109710
|
zfat
|
zinc finger and AT hook domain containing |
| chr18_+_14684115 | 0.23 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr17_+_40989973 | 0.23 |
ENSDART00000160049
|
mrpl33
|
mitochondrial ribosomal protein L33 |
| chr6_-_3998199 | 0.23 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr23_+_25995727 | 0.23 |
ENSDART00000137123
|
wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr1_+_19535144 | 0.23 |
ENSDART00000103089
|
si:dkey-245p14.4
|
si:dkey-245p14.4 |
| chr18_+_20225961 | 0.22 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr6_-_12296170 | 0.22 |
ENSDART00000155685
|
pkp4
|
plakophilin 4 |
| chr12_+_17042754 | 0.22 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr8_-_17997845 | 0.22 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr25_+_15933411 | 0.22 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr24_-_26485098 | 0.22 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr4_+_57580303 | 0.22 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr5_+_36895545 | 0.21 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr4_-_41269844 | 0.21 |
ENSDART00000186177
|
CR388165.2
|
|
| chr21_+_25777425 | 0.21 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr20_-_19858936 | 0.21 |
ENSDART00000161579
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr6_+_22679610 | 0.21 |
ENSDART00000102701
|
zmp:0000000634
|
zmp:0000000634 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0048327 | axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) axial mesoderm structural organization(GO:0048331) mesoderm structural organization(GO:0048338) |
| 0.3 | 0.8 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.2 | 0.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 0.8 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.8 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.2 | 0.6 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.2 | 0.5 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.2 | 0.5 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 0.6 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.9 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 0.7 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.5 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 0.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.6 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.3 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.4 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 0.4 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.8 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.4 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.3 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.3 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.1 | 0.5 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 1.5 | GO:0006949 | syncytium formation by plasma membrane fusion(GO:0000768) syncytium formation(GO:0006949) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.5 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.5 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.3 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.0 | 0.3 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.0 | 0.1 | GO:0001774 | microglial cell activation(GO:0001774) |
| 0.0 | 1.0 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 1.2 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.2 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 1.5 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.1 | GO:0002727 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.0 | 1.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 2.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 0.3 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.0 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.2 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.4 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 1.3 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 2.6 | GO:0042157 | lipoprotein metabolic process(GO:0042157) |
| 0.0 | 0.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.6 | GO:0043523 | regulation of neuron apoptotic process(GO:0043523) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.0 | 0.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.5 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:1901099 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 7.4 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 0.8 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.7 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.3 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.1 | 0.6 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.0 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 1.0 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.0 | GO:0034399 | preribosome, large subunit precursor(GO:0030687) nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 7.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.9 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.6 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.6 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.3 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0000035 | acyl binding(GO:0000035) |
| 0.2 | 0.8 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.1 | 0.6 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.6 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.7 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.6 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.0 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.3 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 7.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.5 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.4 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.5 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.0 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.3 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.6 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.6 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 2.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.8 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.6 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 2.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.3 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |