Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
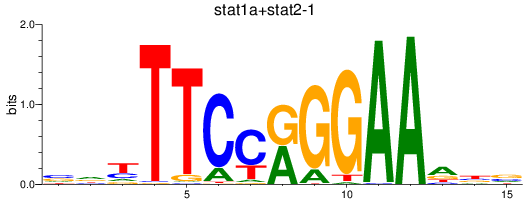
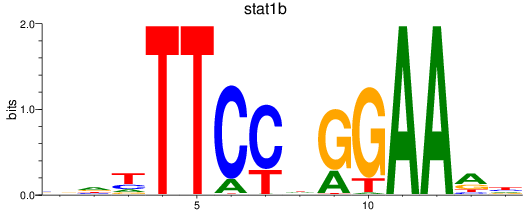
Results for stat1a+stat2-1_stat1b
Z-value: 1.70
Transcription factors associated with stat1a+stat2-1_stat1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat1a
|
ENSDARG00000006266 | signal transducer and activator of transcription 1a |
|
stat2-1
|
ENSDARG00000031647 | signal transducer and activator of transcription 2 |
|
stat1b
|
ENSDARG00000076182 | signal transducer and activator of transcription 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat1a | dr11_v1_chr22_+_12798569_12798569 | 0.95 | 3.4e-10 | Click! |
| stat2 | dr11_v1_chr6_-_39158953_39158953 | 0.66 | 2.0e-03 | Click! |
| stat1b | dr11_v1_chr9_+_41218967_41218974 | 0.59 | 8.2e-03 | Click! |
Activity profile of stat1a+stat2-1_stat1b motif
Sorted Z-values of stat1a+stat2-1_stat1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_61205711 | 12.69 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr12_-_3252263 | 8.28 |
ENSDART00000170632
|
gngt2b
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2b |
| chr16_-_36834505 | 8.00 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr5_-_30620625 | 7.60 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr7_+_73295890 | 7.44 |
ENSDART00000174331
ENSDART00000174250 |
CABZ01083442.1
|
|
| chr20_+_10166297 | 6.98 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr6_+_41503854 | 6.74 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr17_-_6730247 | 6.59 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr21_+_39100289 | 6.19 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr13_+_52061034 | 5.92 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr10_-_11385155 | 5.60 |
ENSDART00000064214
|
plac8.1
|
placenta-specific 8, tandem duplicate 1 |
| chr18_+_49411417 | 5.47 |
ENSDART00000028944
|
LRFN3
|
zmp:0000001073 |
| chr16_-_52879741 | 5.27 |
ENSDART00000166470
|
TPPP
|
tubulin polymerization promoting protein |
| chr24_+_5208171 | 5.17 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr9_-_23922011 | 5.16 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr9_-_6372535 | 5.07 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr9_-_9282519 | 4.70 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr23_+_44611864 | 4.67 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr21_+_45626136 | 4.48 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr18_+_7264961 | 4.43 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr24_+_42948 | 4.27 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr2_-_38125657 | 4.12 |
ENSDART00000143433
|
cbln12
|
cerebellin 12 |
| chr3_+_25154078 | 4.08 |
ENSDART00000156973
|
si:ch211-256m1.8
|
si:ch211-256m1.8 |
| chr14_+_6159162 | 4.01 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr22_+_7480465 | 3.94 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr18_-_16801033 | 3.91 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr2_-_42091926 | 3.78 |
ENSDART00000142663
|
si:dkey-97a13.12
|
si:dkey-97a13.12 |
| chr19_+_48176745 | 3.70 |
ENSDART00000164963
|
prdm1b
|
PR domain containing 1b, with ZNF domain |
| chr1_+_44053641 | 3.69 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr20_+_13969414 | 3.65 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr5_+_23118470 | 3.39 |
ENSDART00000149893
|
nexmifa
|
neurite extension and migration factor a |
| chr4_-_5291256 | 3.35 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr13_+_1535511 | 3.34 |
ENSDART00000159454
|
CABZ01044277.1
|
|
| chr24_+_24923166 | 3.25 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr13_-_37631092 | 3.23 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr12_-_4683325 | 3.22 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr11_-_762721 | 3.03 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr7_-_71837213 | 3.02 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr23_+_20408227 | 3.01 |
ENSDART00000134727
|
si:rp71-17i16.4
|
si:rp71-17i16.4 |
| chr4_-_18436899 | 2.99 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr15_+_16521785 | 2.96 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr7_+_30787903 | 2.92 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr25_-_8160030 | 2.88 |
ENSDART00000067159
|
tph1a
|
tryptophan hydroxylase 1 (tryptophan 5-monooxygenase) a |
| chr22_+_5851122 | 2.77 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr9_-_23922778 | 2.72 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr23_+_45229198 | 2.69 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr17_-_45040813 | 2.66 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr18_+_10820430 | 2.60 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr15_-_29598444 | 2.59 |
ENSDART00000154847
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr18_+_17660402 | 2.59 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr15_-_29598679 | 2.58 |
ENSDART00000155153
|
si:ch211-207n23.2
|
si:ch211-207n23.2 |
| chr11_+_43401592 | 2.58 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr2_+_38556195 | 2.56 |
ENSDART00000138769
|
cdh24b
|
cadherin 24, type 2b |
| chr3_-_44059902 | 2.53 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr3_+_26813058 | 2.52 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr9_-_32813177 | 2.51 |
ENSDART00000012694
|
mxc
|
myxovirus (influenza virus) resistance C |
| chr16_-_17188294 | 2.44 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr20_-_25551676 | 2.40 |
ENSDART00000063081
|
cyp2ad3
|
cytochrome P450, family 2, subfamily AD, polypeptide 3 |
| chr14_-_2206476 | 2.39 |
ENSDART00000081870
|
pcdh2ab6
|
protocadherin 2 alpha b 6 |
| chr12_-_3133483 | 2.38 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr8_-_42677086 | 2.32 |
ENSDART00000191173
|
si:ch73-138n13.1
|
si:ch73-138n13.1 |
| chr10_-_4375190 | 2.31 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr16_-_36099492 | 2.29 |
ENSDART00000180905
|
CU499336.2
|
|
| chr12_+_22607761 | 2.25 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr23_-_39636195 | 2.21 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr1_-_53407087 | 2.21 |
ENSDART00000157910
|
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr9_+_51265283 | 2.20 |
ENSDART00000137426
|
gcgb
|
glucagon b |
| chr8_-_5155545 | 2.13 |
ENSDART00000056885
|
DPYSL2 (1 of many)
|
dihydropyrimidinase like 2 |
| chr2_+_7295515 | 2.10 |
ENSDART00000152987
|
si:dkeyp-106c3.1
|
si:dkeyp-106c3.1 |
| chr16_-_22251414 | 2.08 |
ENSDART00000158500
ENSDART00000179998 |
atp8b2
|
ATPase phospholipid transporting 8B2 |
| chr15_-_2903475 | 2.07 |
ENSDART00000043226
|
guca1c
|
guanylate cyclase activator 1C |
| chr8_-_14052349 | 2.06 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr24_-_5932982 | 2.05 |
ENSDART00000138412
ENSDART00000135124 ENSDART00000007373 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr16_+_45746549 | 2.01 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr11_-_17964525 | 1.99 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr7_-_52842605 | 1.98 |
ENSDART00000083002
|
map1aa
|
microtubule-associated protein 1Aa |
| chr24_+_15897717 | 1.95 |
ENSDART00000105956
|
neto1l
|
neuropilin (NRP) and tolloid (TLL)-like 1, like |
| chr5_-_18897482 | 1.95 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr5_+_68112194 | 1.95 |
ENSDART00000162468
|
CABZ01083937.1
|
|
| chr1_+_37391716 | 1.91 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr21_+_26720803 | 1.89 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr10_+_22134606 | 1.88 |
ENSDART00000155228
|
si:dkey-4c2.11
|
si:dkey-4c2.11 |
| chr3_+_54047342 | 1.87 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr7_+_58686860 | 1.86 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr25_-_20258508 | 1.85 |
ENSDART00000133860
ENSDART00000006840 ENSDART00000173434 |
dnm1l
|
dynamin 1-like |
| chr16_-_45398408 | 1.84 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr11_-_4220167 | 1.82 |
ENSDART00000185406
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr20_-_39273987 | 1.81 |
ENSDART00000127173
|
clu
|
clusterin |
| chr21_+_1568044 | 1.80 |
ENSDART00000193570
ENSDART00000151601 |
rab27b
|
RAB27B, member RAS oncogene family |
| chr14_+_6429399 | 1.79 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr11_+_13224281 | 1.78 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr17_-_2834764 | 1.78 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr1_-_2334254 | 1.76 |
ENSDART00000144500
|
si:ch211-235f1.3
|
si:ch211-235f1.3 |
| chr25_-_17395315 | 1.74 |
ENSDART00000064596
|
cyp2x8
|
cytochrome P450, family 2, subfamily X, polypeptide 8 |
| chr19_+_17385561 | 1.74 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr7_-_38634845 | 1.73 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr23_-_8373676 | 1.72 |
ENSDART00000105135
ENSDART00000158531 |
oprl1
|
opiate receptor-like 1 |
| chr20_+_6493648 | 1.71 |
ENSDART00000099846
|
mep1b
|
meprin A, beta |
| chr8_+_52503340 | 1.70 |
ENSDART00000168838
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr20_+_29586529 | 1.70 |
ENSDART00000178617
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr12_-_26587231 | 1.70 |
ENSDART00000153168
|
slc16a5a
|
solute carrier family 16 (monocarboxylate transporter), member 5a |
| chr16_-_27749172 | 1.66 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr18_+_6479963 | 1.65 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr16_+_32059785 | 1.62 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr6_+_22068589 | 1.61 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr16_+_5774977 | 1.60 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr15_-_31067589 | 1.59 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr24_-_38113208 | 1.57 |
ENSDART00000079003
|
crp
|
c-reactive protein, pentraxin-related |
| chr25_+_6186823 | 1.56 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr5_-_33886495 | 1.55 |
ENSDART00000159058
|
dab2ipb
|
DAB2 interacting protein b |
| chr5_+_1911814 | 1.53 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr6_+_2195625 | 1.53 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr14_+_8328645 | 1.53 |
ENSDART00000127494
|
nrg2b
|
neuregulin 2b |
| chr14_+_6159356 | 1.52 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr8_-_34065573 | 1.52 |
ENSDART00000186946
|
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr1_+_41454210 | 1.52 |
ENSDART00000148251
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr19_-_31042570 | 1.52 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr17_+_27064158 | 1.51 |
ENSDART00000184864
ENSDART00000178734 ENSDART00000129273 |
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr5_+_36768674 | 1.51 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr9_-_54304684 | 1.49 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr13_+_24552254 | 1.48 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr8_+_36948256 | 1.48 |
ENSDART00000140410
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr19_-_20114149 | 1.46 |
ENSDART00000052620
|
npy
|
neuropeptide Y |
| chr11_+_13223625 | 1.45 |
ENSDART00000161275
|
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr24_-_22702017 | 1.43 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr3_+_24511959 | 1.42 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr21_+_15870752 | 1.42 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr9_+_6079528 | 1.41 |
ENSDART00000142167
|
st6gal2a
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2a |
| chr10_+_17776981 | 1.40 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr23_-_1571682 | 1.39 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr6_+_6491013 | 1.36 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr2_+_55199721 | 1.36 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr21_-_30545121 | 1.36 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr24_-_11905911 | 1.35 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr9_+_29585943 | 1.34 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr1_-_21599219 | 1.32 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr18_-_6460102 | 1.31 |
ENSDART00000137037
|
iqsec3b
|
IQ motif and Sec7 domain 3b |
| chr13_-_11644806 | 1.30 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr11_+_43401201 | 1.29 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr4_+_19534833 | 1.29 |
ENSDART00000140028
|
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr4_+_76575585 | 1.27 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr4_+_359970 | 1.26 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr3_-_49110710 | 1.26 |
ENSDART00000160404
|
trim35-12
|
tripartite motif containing 35-12 |
| chr18_-_46135927 | 1.25 |
ENSDART00000134537
ENSDART00000191481 |
SPTBN4
|
si:ch73-262h23.4 |
| chr15_-_30832897 | 1.25 |
ENSDART00000152330
|
msi2b
|
musashi RNA-binding protein 2b |
| chr4_-_9810999 | 1.24 |
ENSDART00000146858
|
si:dkeyp-27e10.3
|
si:dkeyp-27e10.3 |
| chr21_-_22676323 | 1.23 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr12_-_46112892 | 1.22 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr24_+_22056386 | 1.22 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr13_-_37608441 | 1.21 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr1_-_6085750 | 1.20 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr9_-_18877597 | 1.19 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr19_+_41701660 | 1.19 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr8_+_53452681 | 1.19 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr19_-_17385548 | 1.18 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr13_+_33117528 | 1.17 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr7_+_39706004 | 1.17 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr3_+_19621034 | 1.17 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr20_+_34933183 | 1.17 |
ENSDART00000062738
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr5_-_40190949 | 1.16 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr1_-_46343999 | 1.15 |
ENSDART00000145117
ENSDART00000193233 |
atp11a
|
ATPase phospholipid transporting 11A |
| chr13_-_2112008 | 1.15 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr18_+_13792490 | 1.12 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr20_-_25412181 | 1.12 |
ENSDART00000062780
|
itsn2a
|
intersectin 2a |
| chr22_+_20427170 | 1.12 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr9_+_6934491 | 1.10 |
ENSDART00000114323
|
mfsd9
|
major facilitator superfamily domain containing 9 |
| chr7_-_36358303 | 1.10 |
ENSDART00000130028
|
fto
|
fat mass and obesity associated |
| chr24_+_13635387 | 1.10 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr19_-_35596207 | 1.10 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr13_+_40019001 | 1.09 |
ENSDART00000158820
|
golga7bb
|
golgin A7 family, member Bb |
| chr11_-_27962757 | 1.09 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr9_-_5045378 | 1.08 |
ENSDART00000149268
|
nr4a2a
|
nuclear receptor subfamily 4, group A, member 2a |
| chr19_-_6631900 | 1.07 |
ENSDART00000144571
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr10_+_21722892 | 1.07 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr14_+_16345003 | 1.06 |
ENSDART00000003040
ENSDART00000165193 |
itln3
|
intelectin 3 |
| chr19_-_26235219 | 1.05 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr23_+_10469955 | 1.04 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr5_-_42883761 | 1.03 |
ENSDART00000167374
|
BX323596.2
|
|
| chr7_-_36358735 | 1.03 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr15_-_568645 | 1.02 |
ENSDART00000156744
|
cbln18
|
cerebellin 18 |
| chr5_+_20148671 | 1.02 |
ENSDART00000143205
|
svopa
|
SV2 related protein a |
| chr4_+_76637548 | 1.02 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr8_-_31701157 | 0.99 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr13_+_24750078 | 0.99 |
ENSDART00000021053
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr14_+_33329420 | 0.98 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr4_+_76735113 | 0.98 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr7_-_2175181 | 0.97 |
ENSDART00000174011
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr7_+_30867008 | 0.97 |
ENSDART00000193106
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr7_-_20241346 | 0.97 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr17_-_26604946 | 0.97 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr3_-_13461361 | 0.97 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr9_-_12443726 | 0.95 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr14_-_2202652 | 0.94 |
ENSDART00000171316
|
si:dkey-262j3.7
|
si:dkey-262j3.7 |
| chr6_+_18321627 | 0.94 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr1_-_46981134 | 0.94 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr5_+_25952340 | 0.93 |
ENSDART00000147188
|
trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr11_-_18800299 | 0.92 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr6_+_9870192 | 0.91 |
ENSDART00000150894
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr13_-_40238813 | 0.90 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr18_+_3332999 | 0.90 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat1a+stat2-1_stat1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 1.7 | 5.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 1.1 | 4.3 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 1.0 | 3.9 | GO:0043084 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.8 | 3.4 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.8 | 3.2 | GO:0032782 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.8 | 5.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.7 | 2.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.6 | 2.5 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.6 | 4.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.5 | 1.6 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 2.9 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.4 | 1.7 | GO:0015677 | copper ion import(GO:0015677) |
| 0.4 | 4.5 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.4 | 1.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.4 | 1.5 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.4 | 2.8 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.3 | 2.0 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.3 | 1.6 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.3 | 0.9 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.3 | 4.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.3 | 0.9 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.3 | 0.9 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.3 | 0.8 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.3 | 7.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.3 | 2.7 | GO:0072506 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.3 | 1.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.3 | 0.8 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.2 | 1.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.2 | 0.7 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.2 | 0.7 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.2 | 0.9 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.2 | 0.6 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.2 | 2.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.2 | 1.5 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 3.2 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.6 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.2 | 1.8 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.2 | 1.9 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.2 | 5.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 5.2 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.6 | GO:0034653 | vitamin catabolic process(GO:0009111) diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) fat-soluble vitamin catabolic process(GO:0042363) |
| 0.1 | 1.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.7 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.6 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 1.0 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 2.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 3.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.0 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.2 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 2.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 3.0 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 6.7 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.1 | 0.8 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 1.1 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.5 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.1 | 0.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.6 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.1 | 1.6 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.1 | 2.7 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 3.0 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 2.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 0.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.8 | GO:0071222 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.1 | 1.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 1.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 1.8 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 2.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.8 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.2 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.8 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 4.7 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.7 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.0 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.0 | 1.6 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 3.6 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 1.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 2.6 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 5.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.6 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.1 | GO:0038127 | epidermal growth factor receptor signaling pathway(GO:0007173) ERBB signaling pathway(GO:0038127) |
| 0.0 | 1.8 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 4.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 2.2 | GO:0042594 | response to starvation(GO:0042594) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 2.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 1.1 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.7 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 1.0 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.2 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.8 | GO:0048821 | erythrocyte development(GO:0048821) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.8 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.4 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 0.0 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) postsynaptic neurotransmitter receptor internalization(GO:0098884) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 0.6 | GO:0030055 | cell-substrate junction(GO:0030055) |
| 0.6 | 4.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 8.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.3 | 14.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 0.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.2 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 0.6 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.2 | 1.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 2.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 1.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 9.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 3.9 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.7 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 6.2 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.1 | 3.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.8 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 4.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 1.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 3.0 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 4.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 2.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 1.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.9 | GO:0044306 | axon terminus(GO:0043679) neuron projection terminus(GO:0044306) |
| 0.1 | 7.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 1.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.2 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.9 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 1.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 3.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.5 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 4.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.9 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 2.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.3 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 1.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 1.7 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.8 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.8 | 4.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.8 | 3.2 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.7 | 3.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 3.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.6 | 4.3 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.6 | 4.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 2.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.5 | 1.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.5 | 2.1 | GO:0035516 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.5 | 14.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.4 | 2.2 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.4 | 5.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.4 | 1.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.4 | 8.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.4 | 1.6 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.4 | 1.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.4 | 1.8 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 0.9 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 1.9 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.3 | 1.5 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.3 | 7.0 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.3 | 2.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.3 | 1.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 1.0 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.2 | 1.9 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.2 | 1.7 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.2 | 3.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 1.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 3.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 0.5 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.2 | 2.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.6 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 4.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.5 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 1.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 0.7 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.5 | GO:0101006 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.9 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.1 | 2.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 0.7 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.1 | 0.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.7 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.7 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 3.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 2.0 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.1 | 0.8 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 1.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 2.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.4 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 7.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.3 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.1 | 1.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 0.2 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 5.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 4.8 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 4.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 2.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 6.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.0 | 0.3 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.7 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 1.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 1.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 4.1 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 1.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 2.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.9 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 15.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 6.9 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 1.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 3.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 0.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.4 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.0 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 1.6 | GO:0008083 | growth factor activity(GO:0008083) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 6.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 1.5 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 8.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 2.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 1.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 0.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.1 | 3.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 1.9 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 9.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.4 | 7.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.3 | 0.9 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 4.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 7.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 1.8 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 4.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.6 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.7 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |