Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
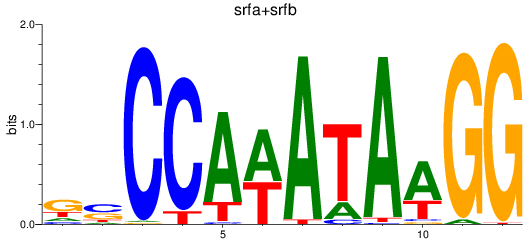
Results for srfa+srfb
Z-value: 2.11
Transcription factors associated with srfa+srfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
srfa
|
ENSDARG00000053918 | serum response factor a |
|
srfb
|
ENSDARG00000102867 | serum response factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| srfb | dr11_v1_chr13_-_51922290_51922290 | -0.77 | 1.3e-04 | Click! |
| srfa | dr11_v1_chr22_+_35089031_35089031 | 0.14 | 5.6e-01 | Click! |
Activity profile of srfa+srfb motif
Sorted Z-values of srfa+srfb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_21336385 | 23.84 |
ENSDART00000054460
|
egr1
|
early growth response 1 |
| chr15_-_551177 | 14.86 |
ENSDART00000066774
ENSDART00000154617 |
tagln
|
transgelin |
| chr8_+_40628926 | 8.85 |
ENSDART00000163598
|
dusp2
|
dual specificity phosphatase 2 |
| chr9_-_44948488 | 8.23 |
ENSDART00000059228
|
vil1
|
villin 1 |
| chr11_+_25459697 | 7.33 |
ENSDART00000161481
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr2_+_58221163 | 7.24 |
ENSDART00000157939
|
FO704813.1
|
|
| chr21_+_30043054 | 6.95 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr17_-_15611744 | 5.37 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr23_+_45579497 | 5.29 |
ENSDART00000110381
|
egr4
|
early growth response 4 |
| chr5_-_32309129 | 5.21 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr5_-_32292965 | 5.17 |
ENSDART00000183522
ENSDART00000131983 |
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr8_+_10561922 | 4.86 |
ENSDART00000133348
|
fam19a5l
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5-like |
| chr11_-_34097418 | 4.80 |
ENSDART00000028301
ENSDART00000138579 |
tdo2b
|
tryptophan 2,3-dioxygenase b |
| chr3_-_35602233 | 4.69 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr12_-_35054354 | 4.63 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr2_+_31804582 | 4.52 |
ENSDART00000086646
|
rnf182
|
ring finger protein 182 |
| chr25_+_33192796 | 4.46 |
ENSDART00000125892
ENSDART00000121680 ENSDART00000014851 |
TPM1 (1 of many)
|
zgc:171719 |
| chr21_+_6780340 | 4.39 |
ENSDART00000139493
ENSDART00000140478 |
olfm1b
|
olfactomedin 1b |
| chr23_+_44611864 | 4.34 |
ENSDART00000145905
ENSDART00000132361 |
eno3
|
enolase 3, (beta, muscle) |
| chr17_-_20979077 | 4.25 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr20_-_46554440 | 4.17 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr12_-_26064480 | 4.01 |
ENSDART00000158215
ENSDART00000171206 ENSDART00000171212 ENSDART00000182956 ENSDART00000186779 |
ldb3b
|
LIM domain binding 3b |
| chr3_+_45401472 | 4.00 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr20_-_29420713 | 3.83 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr3_-_40976463 | 3.79 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr25_+_33192404 | 3.78 |
ENSDART00000193592
|
TPM1 (1 of many)
|
zgc:171719 |
| chr19_-_42424599 | 3.63 |
ENSDART00000077042
|
zgc:153441
|
zgc:153441 |
| chr8_+_50727220 | 3.54 |
ENSDART00000127062
|
egr3
|
early growth response 3 |
| chr7_+_29952169 | 3.54 |
ENSDART00000173540
ENSDART00000173940 ENSDART00000173906 ENSDART00000173772 ENSDART00000173506 ENSDART00000039657 |
tpma
|
alpha-tropomyosin |
| chr3_-_22228602 | 3.49 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr5_+_9428876 | 3.35 |
ENSDART00000081791
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr7_-_18168493 | 3.22 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr23_-_16692312 | 3.06 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr23_+_42813415 | 3.01 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr20_-_40750953 | 2.99 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr12_-_26064105 | 2.92 |
ENSDART00000168825
|
ldb3b
|
LIM domain binding 3b |
| chr4_+_12612723 | 2.82 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr18_+_36769758 | 2.82 |
ENSDART00000180375
ENSDART00000136463 ENSDART00000133487 ENSDART00000130206 |
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr11_+_40657612 | 2.76 |
ENSDART00000183271
|
slc45a1
|
solute carrier family 45, member 1 |
| chr2_+_15100742 | 2.75 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr6_-_55309190 | 2.74 |
ENSDART00000162117
|
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr3_-_33395233 | 2.70 |
ENSDART00000167349
|
si:dkey-283b1.6
|
si:dkey-283b1.6 |
| chr22_+_33362552 | 2.63 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr20_-_14875308 | 2.62 |
ENSDART00000141290
|
dnm3a
|
dynamin 3a |
| chr7_+_29951997 | 2.58 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr13_-_9598320 | 2.49 |
ENSDART00000184613
|
cpxm1a
|
carboxypeptidase X (M14 family), member 1a |
| chr7_-_53117131 | 2.48 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr4_-_16644708 | 2.42 |
ENSDART00000042307
|
sinhcaf
|
SIN3-HDAC complex associated factor |
| chr20_-_47732703 | 2.37 |
ENSDART00000193975
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr18_+_36770166 | 2.35 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr7_-_66868543 | 2.34 |
ENSDART00000149680
|
ampd3a
|
adenosine monophosphate deaminase 3a |
| chr20_+_9211237 | 2.33 |
ENSDART00000139527
|
si:ch211-59d15.4
|
si:ch211-59d15.4 |
| chr15_+_8043751 | 2.33 |
ENSDART00000193701
|
cadm2b
|
cell adhesion molecule 2b |
| chr20_+_13969414 | 2.32 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr23_-_45439903 | 2.30 |
ENSDART00000170729
|
NPNT (1 of many)
|
nephronectin |
| chr17_+_13763425 | 2.28 |
ENSDART00000105175
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr1_-_14332283 | 2.23 |
ENSDART00000090025
|
wfs1a
|
Wolfram syndrome 1a (wolframin) |
| chr7_-_71758307 | 2.19 |
ENSDART00000161067
ENSDART00000165253 |
myom1b
|
myomesin 1b |
| chr4_+_27898833 | 2.14 |
ENSDART00000146099
|
cerk
|
ceramide kinase |
| chr10_+_8101729 | 2.10 |
ENSDART00000138875
ENSDART00000123447 ENSDART00000185333 |
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr10_+_42169982 | 2.06 |
ENSDART00000190905
|
CU467905.1
|
|
| chr12_-_15584479 | 2.05 |
ENSDART00000150134
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr12_-_29624638 | 2.03 |
ENSDART00000126744
|
nrg3b
|
neuregulin 3b |
| chr23_-_44207349 | 2.00 |
ENSDART00000186276
|
zgc:158659
|
zgc:158659 |
| chr1_+_50976975 | 1.98 |
ENSDART00000022290
ENSDART00000140982 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr2_-_37477654 | 1.96 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr1_+_9071324 | 1.95 |
ENSDART00000184194
|
BX004779.2
|
|
| chr11_-_24016761 | 1.95 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr3_-_40945710 | 1.95 |
ENSDART00000138719
ENSDART00000102416 |
cyp3c4
|
cytochrome P450, family 3, subfamily C, polypeptide 4 |
| chr22_-_29336268 | 1.93 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr18_-_14941840 | 1.93 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr11_-_2250767 | 1.88 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr4_+_5180650 | 1.88 |
ENSDART00000067390
|
fgf6b
|
fibroblast growth factor 6b |
| chr24_+_31277360 | 1.87 |
ENSDART00000165993
|
f3a
|
coagulation factor IIIa |
| chr12_-_20120702 | 1.85 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr12_-_35095414 | 1.83 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr23_-_35064785 | 1.76 |
ENSDART00000172240
|
BX294434.1
|
|
| chr20_+_53577502 | 1.76 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr3_+_32553714 | 1.75 |
ENSDART00000165638
|
pax10
|
paired box 10 |
| chr12_+_17100021 | 1.69 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr13_+_24279021 | 1.65 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr20_+_40150612 | 1.65 |
ENSDART00000143680
ENSDART00000109681 ENSDART00000101041 ENSDART00000121818 |
trdn
|
triadin |
| chr2_+_30916188 | 1.63 |
ENSDART00000137012
|
myom1a
|
myomesin 1a (skelemin) |
| chr10_-_7892192 | 1.62 |
ENSDART00000145480
|
smtna
|
smoothelin a |
| chr20_-_2949028 | 1.62 |
ENSDART00000104667
ENSDART00000193151 ENSDART00000131946 |
cdk19
|
cyclin-dependent kinase 19 |
| chr10_-_7857494 | 1.60 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr12_+_42436328 | 1.60 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr19_+_43715911 | 1.59 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr6_-_13498745 | 1.55 |
ENSDART00000027684
ENSDART00000189438 |
mylkb
|
myosin light chain kinase b |
| chr1_+_44173245 | 1.55 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr10_-_15910974 | 1.54 |
ENSDART00000148169
|
pip5k1ba
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta a |
| chr7_-_22324843 | 1.54 |
ENSDART00000187408
|
CU571061.1
|
|
| chr4_-_6623645 | 1.49 |
ENSDART00000060861
|
foxp2
|
forkhead box P2 |
| chr13_-_40398556 | 1.46 |
ENSDART00000191921
|
nkx3.3
|
NK3 homeobox 3 |
| chr18_+_45504362 | 1.45 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr11_+_37178271 | 1.43 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr2_+_38264964 | 1.42 |
ENSDART00000182068
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr13_+_23132666 | 1.39 |
ENSDART00000164639
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr1_+_9994811 | 1.37 |
ENSDART00000143719
ENSDART00000110749 |
si:dkeyp-75b4.10
|
si:dkeyp-75b4.10 |
| chr13_-_18695289 | 1.37 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr11_-_472547 | 1.34 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr10_-_29816467 | 1.33 |
ENSDART00000055913
|
hist2h2l
|
histone 2, H2, like |
| chr21_+_8533533 | 1.28 |
ENSDART00000077924
|
FO834888.1
|
|
| chr3_-_37351225 | 1.27 |
ENSDART00000174685
|
si:ch211-278a6.1
|
si:ch211-278a6.1 |
| chr5_-_55395964 | 1.25 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr12_-_20373058 | 1.24 |
ENSDART00000066382
|
aqp8a.1
|
aquaporin 8a, tandem duplicate 1 |
| chr2_-_24269911 | 1.23 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr10_-_690072 | 1.22 |
ENSDART00000164871
ENSDART00000142833 |
glis3
|
GLIS family zinc finger 3 |
| chr1_+_57331813 | 1.21 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr7_+_56651759 | 1.20 |
ENSDART00000073600
|
kcng4b
|
potassium voltage-gated channel, subfamily G, member 4b |
| chr14_+_48964628 | 1.19 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr22_-_11493236 | 1.15 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr3_-_18410968 | 1.10 |
ENSDART00000041842
|
ndufb10
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10 |
| chr14_+_17137023 | 1.10 |
ENSDART00000080712
|
slc43a3b
|
solute carrier family 43, member 3b |
| chr5_-_41845116 | 1.09 |
ENSDART00000112382
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr20_+_1996202 | 1.08 |
ENSDART00000184143
|
CABZ01092781.1
|
|
| chr8_+_26268726 | 1.05 |
ENSDART00000180883
|
slc26a6
|
solute carrier family 26, member 6 |
| chr21_+_4509483 | 1.03 |
ENSDART00000025612
|
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr1_-_37383741 | 0.99 |
ENSDART00000193155
ENSDART00000191887 ENSDART00000189077 |
scpp1
|
secretory calcium-binding phosphoprotein 1 |
| chr8_+_36942262 | 0.97 |
ENSDART00000188173
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr12_+_35654749 | 0.95 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr25_-_19031019 | 0.94 |
ENSDART00000191707
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr21_+_4508959 | 0.93 |
ENSDART00000140432
ENSDART00000147187 ENSDART00000148910 |
phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr17_-_25326296 | 0.90 |
ENSDART00000168822
|
pabpc4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr7_+_47243564 | 0.86 |
ENSDART00000098942
ENSDART00000162237 |
znf507
|
zinc finger protein 507 |
| chr13_+_8840772 | 0.84 |
ENSDART00000059321
|
epcam
|
epithelial cell adhesion molecule |
| chr23_-_37514493 | 0.77 |
ENSDART00000083267
|
dnajc16
|
DnaJ (Hsp40) homolog, subfamily C, member 16 |
| chr2_-_24270062 | 0.77 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr3_-_32898626 | 0.75 |
ENSDART00000103201
|
kat7a
|
K(lysine) acetyltransferase 7a |
| chr9_-_202805 | 0.74 |
ENSDART00000182260
|
CABZ01067557.1
|
|
| chr17_+_45454943 | 0.74 |
ENSDART00000074838
|
kcnk3b
|
potassium channel, subfamily K, member 3b |
| chr3_-_2592350 | 0.70 |
ENSDART00000192325
|
si:dkey-217f16.5
|
si:dkey-217f16.5 |
| chr6_+_58915889 | 0.67 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr4_-_22310956 | 0.63 |
ENSDART00000162585
|
hcls1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr21_+_25765734 | 0.62 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr3_+_23707691 | 0.62 |
ENSDART00000025449
|
hoxb5a
|
homeobox B5a |
| chr15_-_42760110 | 0.61 |
ENSDART00000152490
|
si:ch211-181d7.3
|
si:ch211-181d7.3 |
| chr7_+_24522308 | 0.61 |
ENSDART00000173542
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr7_-_71758613 | 0.61 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr13_-_50002852 | 0.58 |
ENSDART00000099439
|
lyst
|
lysosomal trafficking regulator |
| chr25_-_893464 | 0.57 |
ENSDART00000159321
|
znf609a
|
zinc finger protein 609a |
| chr10_+_29771256 | 0.57 |
ENSDART00000193195
|
hyou1
|
hypoxia up-regulated 1 |
| chr20_+_20186706 | 0.56 |
ENSDART00000002507
|
rhoj
|
ras homolog family member J |
| chr16_-_31675669 | 0.56 |
ENSDART00000168848
ENSDART00000158331 |
c1r
|
complement component 1, r subcomponent |
| chr21_-_25555355 | 0.55 |
ENSDART00000144228
|
si:dkey-17e16.9
|
si:dkey-17e16.9 |
| chr11_+_6281647 | 0.54 |
ENSDART00000002459
|
ctns
|
cystinosin, lysosomal cystine transporter |
| chr22_-_36856405 | 0.53 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr1_+_55293424 | 0.53 |
ENSDART00000152464
|
zgc:172106
|
zgc:172106 |
| chr2_+_47718605 | 0.53 |
ENSDART00000189180
ENSDART00000148824 |
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr12_+_20693743 | 0.52 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr16_-_31754102 | 0.49 |
ENSDART00000185043
|
ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr18_+_50669456 | 0.48 |
ENSDART00000163005
|
si:dkey-151j17.4
|
si:dkey-151j17.4 |
| chr3_+_1724941 | 0.44 |
ENSDART00000193402
|
BX321875.2
|
|
| chr21_+_21811867 | 0.42 |
ENSDART00000185752
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr20_+_46572550 | 0.40 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr14_+_164556 | 0.40 |
ENSDART00000185606
|
WDR1
|
WD repeat domain 1 |
| chr14_-_32824380 | 0.32 |
ENSDART00000172791
ENSDART00000105745 |
inppl1b
|
inositol polyphosphate phosphatase-like 1b |
| chr21_-_31286428 | 0.30 |
ENSDART00000185334
|
ca4c
|
carbonic anhydrase IV c |
| chr14_-_25309058 | 0.30 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr1_+_6307305 | 0.28 |
ENSDART00000183870
|
LO018629.1
|
|
| chr23_-_44466257 | 0.28 |
ENSDART00000150126
|
si:ch1073-228j22.2
|
si:ch1073-228j22.2 |
| chr13_-_25303303 | 0.27 |
ENSDART00000133153
|
plaua
|
plasminogen activator, urokinase a |
| chr6_-_41135215 | 0.27 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr18_-_26675699 | 0.27 |
ENSDART00000113280
|
FRMD5
|
si:ch211-69m14.1 |
| chr4_-_17642168 | 0.25 |
ENSDART00000007030
|
klhl42
|
kelch-like family, member 42 |
| chr22_-_17458070 | 0.25 |
ENSDART00000139658
|
si:ch211-197g15.10
|
si:ch211-197g15.10 |
| chr10_-_373575 | 0.25 |
ENSDART00000114487
|
DMPK
|
DM1 protein kinase |
| chr1_-_6494384 | 0.23 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr16_+_23816933 | 0.23 |
ENSDART00000183428
|
si:dkey-7f3.9
|
si:dkey-7f3.9 |
| chr3_-_56896702 | 0.21 |
ENSDART00000023265
|
ush1ga
|
Usher syndrome 1Ga (autosomal recessive) |
| chr4_-_49133107 | 0.19 |
ENSDART00000150806
|
znf1146
|
zinc finger protein 1146 |
| chr14_-_10492072 | 0.14 |
ENSDART00000182525
|
lpar4
|
lysophosphatidic acid receptor 4 |
| chr19_+_348729 | 0.13 |
ENSDART00000114284
|
mcl1a
|
MCL1, BCL2 family apoptosis regulator a |
| chr25_-_19486399 | 0.13 |
ENSDART00000155076
ENSDART00000156016 |
zgc:193812
|
zgc:193812 |
| chr22_-_382955 | 0.12 |
ENSDART00000082406
|
ora3
|
olfactory receptor class A related 3 |
| chr1_+_44173506 | 0.10 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr23_+_36118738 | 0.09 |
ENSDART00000139319
|
hoxc5a
|
homeobox C5a |
| chr15_-_39945036 | 0.07 |
ENSDART00000192481
|
msh5
|
mutS homolog 5 |
| chr13_-_9070754 | 0.06 |
ENSDART00000143783
ENSDART00000102121 ENSDART00000140820 ENSDART00000184210 |
si:dkey-112g5.12
|
si:dkey-112g5.12 |
| chr1_+_8492379 | 0.05 |
ENSDART00000184518
|
myo15ab
|
myosin XVAb |
| chr10_-_32920690 | 0.02 |
ENSDART00000136245
|
cux1a
|
cut-like homeobox 1a |
| chr1_+_52560549 | 0.01 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr16_-_12496632 | 0.00 |
ENSDART00000019941
|
slc2a3b
|
solute carrier family 2 (facilitated glucose transporter), member 3b |
| chr8_+_14986833 | 0.00 |
ENSDART00000171867
ENSDART00000191374 |
fnbp1l
|
formin binding protein 1-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of srfa+srfb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.6 | 3.2 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.6 | 4.8 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.6 | 23.8 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.5 | 4.3 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.5 | 7.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.4 | 2.5 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.4 | 4.8 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.4 | 1.2 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.4 | 2.0 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.4 | 1.8 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.3 | 2.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.3 | 4.2 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 3.2 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 1.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.2 | 1.6 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 0.5 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 1.9 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 1.9 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.2 | 1.0 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 0.7 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 2.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 2.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.4 | GO:0001562 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 1.2 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.1 | 5.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 3.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 1.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 1.3 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 1.9 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.5 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 2.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.1 | 1.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.1 | 4.7 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.1 | 0.9 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.1 | 6.1 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.1 | 2.2 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.5 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.4 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.3 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 2.4 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.7 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.7 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 2.1 | GO:0006672 | ceramide metabolic process(GO:0006672) |
| 0.0 | 1.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 1.9 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 3.0 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 1.9 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 1.4 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 1.3 | GO:0046474 | glycerophospholipid biosynthetic process(GO:0046474) |
| 0.0 | 0.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 1.9 | GO:0046488 | phosphatidylinositol metabolic process(GO:0046488) |
| 0.0 | 0.6 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 4.0 | GO:0006887 | exocytosis(GO:0006887) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 8.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.5 | 4.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.3 | 2.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 4.4 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 2.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 0.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 8.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 2.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 2.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 5.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 13.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 4.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.1 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.2 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 2.6 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 1.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 13.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 3.6 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 1.2 | 7.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 1.0 | 23.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.7 | 2.1 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.5 | 0.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.5 | 4.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 7.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 2.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.4 | 2.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.4 | 1.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.3 | 6.9 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.3 | 2.8 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 0.7 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.2 | 1.4 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 4.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 1.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 2.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.2 | 2.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 1.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 1.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 7.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.6 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 1.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.1 | 5.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.4 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 2.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 2.0 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 5.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 1.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.9 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 3.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 2.1 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 3.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.1 | 19.3 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 2.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 4.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 2.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 1.2 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 2.5 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 3.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.9 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 32.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.3 | 2.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 2.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 15.2 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.1 | 3.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 5.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 1.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.1 | 0.5 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 2.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 7.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.8 | 24.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.8 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 4.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 7.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.1 | 2.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 1.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 4.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.6 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |