Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
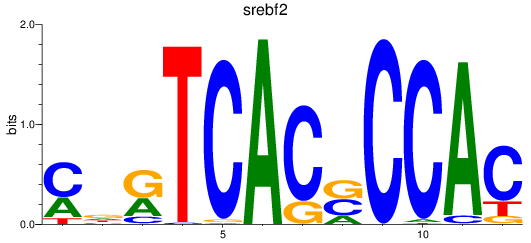
Results for srebf2
Z-value: 0.30
Transcription factors associated with srebf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
srebf2
|
ENSDARG00000063438 | sterol regulatory element binding transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| srebf2 | dr11_v1_chr3_+_1107102_1107102 | 0.59 | 8.5e-03 | Click! |
Activity profile of srebf2 motif
Sorted Z-values of srebf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_992458 | 1.07 |
ENSDART00000114655
|
BFSP1
|
beaded filament structural protein 1 |
| chr7_+_22616212 | 0.67 |
ENSDART00000052844
|
cldn7a
|
claudin 7a |
| chr19_-_10196370 | 0.65 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr2_+_24203229 | 0.49 |
ENSDART00000138088
|
map4l
|
microtubule associated protein 4 like |
| chr10_-_5053589 | 0.49 |
ENSDART00000193579
|
tmem150c
|
transmembrane protein 150C |
| chr9_+_54417141 | 0.47 |
ENSDART00000056810
|
drd1b
|
dopamine receptor D1b |
| chr21_-_19006631 | 0.46 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr22_+_17286841 | 0.44 |
ENSDART00000133176
|
FAM163A
|
si:dkey-171o17.8 |
| chr10_-_31782616 | 0.42 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr15_+_30157602 | 0.42 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr4_+_19535946 | 0.41 |
ENSDART00000192342
ENSDART00000183740 ENSDART00000180812 ENSDART00000180017 |
lrrc4.1
|
leucine rich repeat containing 4.1 |
| chr3_+_19299309 | 0.41 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr6_-_15101477 | 0.37 |
ENSDART00000187713
ENSDART00000124132 |
fhl2b
|
four and a half LIM domains 2b |
| chr16_+_41874001 | 0.37 |
ENSDART00000161278
|
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr16_+_41873708 | 0.34 |
ENSDART00000084631
ENSDART00000084639 ENSDART00000058611 |
scn1ba
|
sodium channel, voltage-gated, type I, beta a |
| chr7_-_30082931 | 0.32 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr22_-_13466246 | 0.31 |
ENSDART00000134035
|
cntnap5b
|
contactin associated protein-like 5b |
| chr10_-_35220285 | 0.30 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr11_+_14280598 | 0.30 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr21_+_28445052 | 0.30 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr13_+_33703784 | 0.30 |
ENSDART00000173361
|
macrod2
|
MACRO domain containing 2 |
| chr21_-_43550120 | 0.30 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr12_+_33395748 | 0.29 |
ENSDART00000129458
|
fasn
|
fatty acid synthase |
| chr7_-_71486162 | 0.28 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr17_-_48915427 | 0.28 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr9_-_4075620 | 0.28 |
ENSDART00000046903
|
mylka
|
myosin, light chain kinase a |
| chr18_+_14342326 | 0.27 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr14_-_33894915 | 0.27 |
ENSDART00000143290
|
urp1
|
urotensin-related peptide 1 |
| chr20_-_7176809 | 0.27 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr1_-_9940494 | 0.26 |
ENSDART00000138726
|
tmem8a
|
transmembrane protein 8A |
| chr20_-_27311675 | 0.26 |
ENSDART00000026088
ENSDART00000148361 |
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr18_-_21271373 | 0.25 |
ENSDART00000060001
|
pnp6
|
purine nucleoside phosphorylase 6 |
| chr12_-_23009312 | 0.23 |
ENSDART00000111801
|
mkxa
|
mohawk homeobox a |
| chr3_-_61099004 | 0.23 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr8_+_22916536 | 0.22 |
ENSDART00000136403
|
cacna1fb
|
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr18_-_39188664 | 0.22 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr5_-_12524996 | 0.20 |
ENSDART00000142258
|
si:ch73-263f13.1
|
si:ch73-263f13.1 |
| chr23_-_27694960 | 0.20 |
ENSDART00000135129
|
IKZF4
|
si:dkey-166n8.9 |
| chr3_+_47245764 | 0.19 |
ENSDART00000193581
|
tnfsf14
|
TNF superfamily member 14 |
| chr6_+_10094061 | 0.19 |
ENSDART00000150998
ENSDART00000162236 |
atp7b
|
ATPase copper transporting beta |
| chr5_-_69948099 | 0.18 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr22_-_13165186 | 0.18 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr12_+_2660733 | 0.18 |
ENSDART00000152235
|
irbpl
|
interphotoreceptor retinoid-binding protein like |
| chr5_+_45976130 | 0.17 |
ENSDART00000175670
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr1_+_9557212 | 0.16 |
ENSDART00000111131
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr2_+_10127762 | 0.16 |
ENSDART00000100726
|
insl5b
|
insulin-like 5b |
| chr24_-_28229618 | 0.16 |
ENSDART00000145290
|
bcl2a
|
BCL2, apoptosis regulator a |
| chr3_-_34296361 | 0.16 |
ENSDART00000181485
ENSDART00000113726 |
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr11_+_14281100 | 0.15 |
ENSDART00000193664
ENSDART00000161597 |
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr6_-_15639087 | 0.15 |
ENSDART00000128940
ENSDART00000183092 |
mlpha
|
melanophilin a |
| chr6_+_102506 | 0.14 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr3_-_45308394 | 0.13 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr1_+_41690402 | 0.13 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr6_-_19351495 | 0.13 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr24_+_40860320 | 0.12 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr14_-_46117345 | 0.12 |
ENSDART00000172166
|
NDUFC1
|
si:ch211-235e9.6 |
| chr3_+_12755535 | 0.11 |
ENSDART00000161286
|
cyp2k17
|
cytochrome P450, family 2, subfamily K, polypeptide17 |
| chr17_-_7436766 | 0.10 |
ENSDART00000162002
|
grm1b
|
glutamate receptor, metabotropic 1b |
| chr21_-_1742159 | 0.10 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr9_-_30363770 | 0.10 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr12_-_34459601 | 0.10 |
ENSDART00000111717
|
fscn2b
|
fascin actin-bundling protein 2b, retinal |
| chr22_+_344763 | 0.10 |
ENSDART00000181934
|
CU914780.1
|
|
| chr1_-_30689004 | 0.08 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr18_+_39649825 | 0.07 |
ENSDART00000087960
|
gldn
|
gliomedin |
| chr24_+_9590188 | 0.06 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr1_-_20826023 | 0.06 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr5_+_37032038 | 0.05 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr1_+_12178215 | 0.05 |
ENSDART00000090380
|
stra6l
|
STRA6-like |
| chr23_-_20258490 | 0.05 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr18_-_27628784 | 0.05 |
ENSDART00000180885
|
BX005470.1
|
|
| chr4_+_41311345 | 0.05 |
ENSDART00000151905
|
si:dkey-16p19.8
|
si:dkey-16p19.8 |
| chr22_-_6959769 | 0.05 |
ENSDART00000105866
|
si:ch73-213k20.5
|
si:ch73-213k20.5 |
| chr3_+_1107102 | 0.04 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr11_-_3334248 | 0.04 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr5_-_15321451 | 0.04 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr8_-_26961779 | 0.03 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr16_+_30316177 | 0.03 |
ENSDART00000109505
|
tmem79b
|
transmembrane protein 79b |
| chr7_-_30932094 | 0.03 |
ENSDART00000057677
|
terb2
|
telomere repeat binding bouquet formation protein 2 |
| chr9_-_38579758 | 0.03 |
ENSDART00000131738
|
si:dkey-101k6.5
|
si:dkey-101k6.5 |
| chr24_-_37326236 | 0.03 |
ENSDART00000003789
|
pdpk1b
|
3-phosphoinositide dependent protein kinase 1b |
| chr13_+_4409294 | 0.02 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr15_+_22333778 | 0.01 |
ENSDART00000114937
|
pde9al
|
phosphodiesterase 9A like |
| chr22_-_8931093 | 0.01 |
ENSDART00000181813
|
FO393424.3
|
|
| chr3_+_14558188 | 0.01 |
ENSDART00000143904
|
epor
|
erythropoietin receptor |
| chr18_+_8231138 | 0.01 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr24_-_40860603 | 0.00 |
ENSDART00000188032
|
CU633479.7
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of srebf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.3 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 0.3 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.1 | 0.4 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.5 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.2 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.7 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.1 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.1 | 0.4 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.3 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.5 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.5 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |