Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
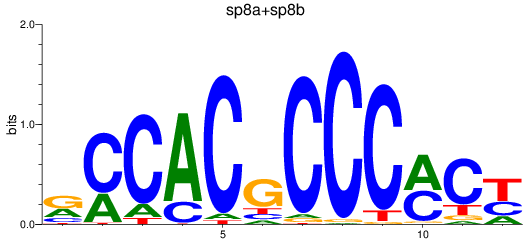
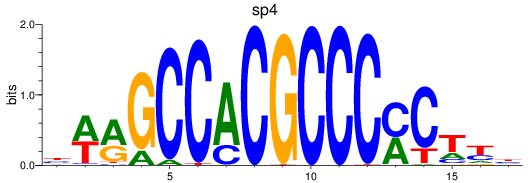
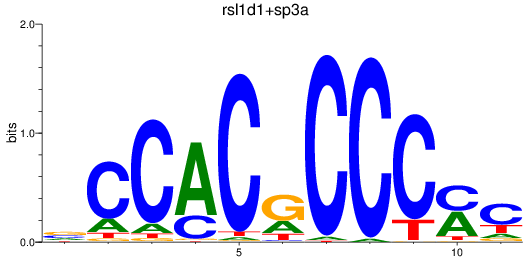
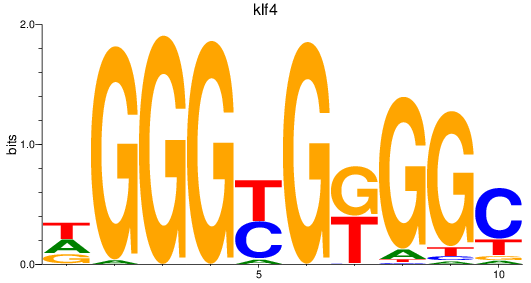
Results for sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
Z-value: 2.05
Transcription factors associated with sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp8a
|
ENSDARG00000011870 | sp8 transcription factor a |
|
sp8b
|
ENSDARG00000056666 | sp8 transcription factor b |
|
sp4
|
ENSDARG00000005186 | sp4 transcription factor |
|
sp3a
|
ENSDARG00000001549 | sp3a transcription factor |
|
rsl1d1
|
ENSDARG00000005846 | Sp5 transcription factor b |
|
klf4
|
ENSDARG00000079922 | Kruppel-like factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp8a | dr11_v1_chr19_-_2317558_2317558 | -0.89 | 2.5e-07 | Click! |
| sp4 | dr11_v1_chr19_+_2364552_2364552 | -0.78 | 9.0e-05 | Click! |
| sp8b | dr11_v1_chr16_+_19537073_19537073 | -0.66 | 2.1e-03 | Click! |
| sp3a | dr11_v1_chr9_+_2762270_2762415 | 0.64 | 3.1e-03 | Click! |
| FO704755.1 | dr11_v1_chr6_+_3730843_3730843 | -0.53 | 2.1e-02 | Click! |
| klf4 | dr11_v1_chr21_-_435466_435466 | 0.33 | 1.6e-01 | Click! |
Activity profile of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4 motif
Sorted Z-values of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_58221163 | 12.59 |
ENSDART00000157939
|
FO704813.1
|
|
| chr16_+_53455638 | 12.26 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr3_-_62380146 | 9.05 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr5_+_67812062 | 8.36 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr17_+_27456804 | 8.21 |
ENSDART00000017756
ENSDART00000181461 ENSDART00000180178 |
ctsl.1
|
cathepsin L.1 |
| chr5_-_50992690 | 8.16 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr2_+_413370 | 7.44 |
ENSDART00000122138
|
mylk4a
|
myosin light chain kinase family, member 4a |
| chr3_+_5575313 | 7.16 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr5_-_69482891 | 6.88 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr1_+_54677173 | 6.78 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr3_+_19299309 | 6.06 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr11_-_4235811 | 5.86 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr22_+_37874691 | 5.73 |
ENSDART00000028565
|
ahsg1
|
alpha-2-HS-glycoprotein 1 |
| chr10_+_1668106 | 5.46 |
ENSDART00000142278
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr6_-_607063 | 5.41 |
ENSDART00000189900
|
lgals2b
|
lectin, galactoside-binding, soluble, 2b |
| chr11_+_1796426 | 5.39 |
ENSDART00000173330
|
lrp1aa
|
low density lipoprotein receptor-related protein 1Aa |
| chr9_+_42095220 | 5.32 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr21_-_39670375 | 5.20 |
ENSDART00000151567
|
sgk494b
|
uncharacterized serine/threonine-protein kinase SgK494b |
| chr10_-_22831611 | 4.98 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr5_-_13086616 | 4.89 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr18_-_40684756 | 4.88 |
ENSDART00000113799
ENSDART00000139042 |
si:ch211-132b12.7
|
si:ch211-132b12.7 |
| chr8_+_1187928 | 4.81 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr8_+_23213320 | 4.65 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr10_+_439692 | 4.56 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr8_-_410728 | 4.48 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr9_-_11263228 | 4.41 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr11_+_6819050 | 4.40 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr5_-_1047222 | 4.40 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr10_+_1638876 | 4.39 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr6_-_16406210 | 4.25 |
ENSDART00000012023
|
faimb
|
Fas apoptotic inhibitory molecule b |
| chr8_+_1082100 | 4.22 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr7_-_6592142 | 4.21 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr25_+_35375848 | 4.16 |
ENSDART00000155721
|
ano3
|
anoctamin 3 |
| chr24_-_14712427 | 4.15 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr18_-_8312848 | 4.13 |
ENSDART00000092033
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr10_+_5135842 | 4.11 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr1_-_59176949 | 4.07 |
ENSDART00000128742
|
CABZ01118678.1
|
|
| chr5_-_46273938 | 3.99 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr20_+_13175379 | 3.98 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr18_-_8313686 | 3.88 |
ENSDART00000182187
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr23_-_41762797 | 3.87 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr22_-_6562618 | 3.86 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr7_+_67325933 | 3.82 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr1_+_9004719 | 3.77 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr2_+_7192966 | 3.77 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr19_+_9305964 | 3.67 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr2_+_24177190 | 3.64 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr18_+_507835 | 3.63 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr3_+_49074008 | 3.58 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr16_-_20707742 | 3.57 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr3_+_11568523 | 3.57 |
ENSDART00000179668
|
CABZ01087224.1
|
|
| chr23_-_30960506 | 3.55 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr3_+_7617353 | 3.41 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr5_+_4332220 | 3.41 |
ENSDART00000051699
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr15_+_24388782 | 3.38 |
ENSDART00000191661
ENSDART00000179995 ENSDART00000111226 |
sez6b
|
seizure related 6 homolog b |
| chr17_-_4318393 | 3.34 |
ENSDART00000167995
ENSDART00000153824 |
napba
|
N-ethylmaleimide-sensitive factor attachment protein, beta a |
| chr22_-_607812 | 3.32 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr25_+_37443194 | 3.32 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr5_-_12031174 | 3.32 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr16_-_6821927 | 3.29 |
ENSDART00000149070
ENSDART00000149570 |
mbpb
|
myelin basic protein b |
| chr21_+_1382078 | 3.27 |
ENSDART00000188463
|
TCF4
|
transcription factor 4 |
| chr6_+_34511886 | 3.27 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr24_-_31439841 | 3.26 |
ENSDART00000169952
|
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr13_-_18691041 | 3.24 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr5_-_68022631 | 3.17 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr3_-_11624694 | 3.12 |
ENSDART00000127157
|
hlfa
|
hepatic leukemia factor a |
| chr6_+_40661703 | 3.11 |
ENSDART00000142492
|
eno1b
|
enolase 1b, (alpha) |
| chr11_-_103136 | 3.09 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr6_-_7776612 | 3.08 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr13_+_1100197 | 3.06 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr10_+_41668483 | 3.05 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr15_-_30816370 | 3.02 |
ENSDART00000142982
ENSDART00000050649 ENSDART00000136901 ENSDART00000100194 |
msi2b
|
musashi RNA-binding protein 2b |
| chr21_-_43398122 | 2.98 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr23_-_41762956 | 2.97 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr23_+_14590767 | 2.97 |
ENSDART00000143675
|
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr22_-_6801876 | 2.95 |
ENSDART00000135726
|
si:ch1073-188e1.1
|
si:ch1073-188e1.1 |
| chr15_-_43164591 | 2.93 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr19_+_1606185 | 2.93 |
ENSDART00000092183
|
lrrc3b
|
leucine rich repeat containing 3B |
| chr23_+_45822935 | 2.91 |
ENSDART00000161892
|
vdra
|
vitamin D receptor a |
| chr23_+_28693278 | 2.87 |
ENSDART00000078148
|
smc1a
|
structural maintenance of chromosomes 1A |
| chr21_-_43079161 | 2.87 |
ENSDART00000144151
|
jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr2_-_48966431 | 2.86 |
ENSDART00000147948
|
kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr3_-_16227683 | 2.84 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr20_-_25518488 | 2.84 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr4_+_57093908 | 2.81 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr25_+_24250247 | 2.81 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr3_+_11548516 | 2.80 |
ENSDART00000059117
|
mmd
|
monocyte to macrophage differentiation-associated |
| chr2_-_34555945 | 2.80 |
ENSDART00000056671
|
brinp2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 2 |
| chr19_+_47299212 | 2.76 |
ENSDART00000158262
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr15_-_47956388 | 2.75 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr20_+_42565049 | 2.75 |
ENSDART00000061101
|
igf2r
|
insulin-like growth factor 2 receptor |
| chr9_-_54001502 | 2.74 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr2_+_38924975 | 2.74 |
ENSDART00000109219
|
rem2
|
RAS (RAD and GEM)-like GTP binding 2 |
| chr5_-_8483457 | 2.71 |
ENSDART00000171886
ENSDART00000159477 ENSDART00000157423 |
lifra
|
leukemia inhibitory factor receptor alpha a |
| chr25_+_4541211 | 2.71 |
ENSDART00000129978
|
pnpla2
|
patatin-like phospholipase domain containing 2 |
| chr8_+_53051701 | 2.69 |
ENSDART00000131514
|
nadka
|
NAD kinase a |
| chr8_-_4100365 | 2.65 |
ENSDART00000142846
|
cux2b
|
cut-like homeobox 2b |
| chr8_-_53490376 | 2.65 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr21_+_45386033 | 2.64 |
ENSDART00000151773
|
jade2
|
jade family PHD finger 2 |
| chr11_+_39959039 | 2.64 |
ENSDART00000024304
|
per3
|
period circadian clock 3 |
| chr24_-_14711597 | 2.63 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr5_+_11812089 | 2.61 |
ENSDART00000111359
|
fbxo21
|
F-box protein 21 |
| chr25_-_19655820 | 2.60 |
ENSDART00000149585
ENSDART00000104353 |
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr19_-_15397946 | 2.60 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr23_+_14590483 | 2.59 |
ENSDART00000088359
ENSDART00000184868 |
si:rp71-79p20.2
|
si:rp71-79p20.2 |
| chr6_+_47424518 | 2.59 |
ENSDART00000165939
|
si:ch211-286o17.1
|
si:ch211-286o17.1 |
| chr8_+_52637507 | 2.59 |
ENSDART00000163830
|
si:dkey-90l8.3
|
si:dkey-90l8.3 |
| chr8_+_7740004 | 2.56 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr20_+_46741074 | 2.54 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr10_-_41352502 | 2.54 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr7_-_24520866 | 2.52 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr19_-_27858033 | 2.49 |
ENSDART00000103898
ENSDART00000144884 |
srd5a1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr2_-_37478418 | 2.48 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr6_-_60104628 | 2.48 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr13_+_47821524 | 2.44 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr10_+_4875262 | 2.42 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr6_-_52156427 | 2.41 |
ENSDART00000082821
|
rims4
|
regulating synaptic membrane exocytosis 4 |
| chr15_+_47418565 | 2.41 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
| chr4_-_22519516 | 2.41 |
ENSDART00000130409
ENSDART00000186258 ENSDART00000002851 ENSDART00000123801 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr8_+_6954984 | 2.40 |
ENSDART00000145610
|
si:ch211-255g12.6
|
si:ch211-255g12.6 |
| chr2_-_48153945 | 2.40 |
ENSDART00000146553
|
pfkpb
|
phosphofructokinase, platelet b |
| chr20_-_1635922 | 2.40 |
ENSDART00000181502
|
CR846082.1
|
|
| chr8_+_2656231 | 2.39 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr19_-_6193448 | 2.37 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr2_+_48073972 | 2.36 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr14_+_22172047 | 2.35 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr1_+_7189856 | 2.29 |
ENSDART00000092114
|
si:ch73-383l1.1
|
si:ch73-383l1.1 |
| chr3_+_59117136 | 2.27 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr24_-_38644937 | 2.25 |
ENSDART00000170194
|
slc6a16b
|
solute carrier family 6, member 16b |
| chr25_-_20268027 | 2.23 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr22_-_29336268 | 2.23 |
ENSDART00000132776
ENSDART00000186351 ENSDART00000121599 |
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr21_+_8427059 | 2.22 |
ENSDART00000143151
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr24_-_29777413 | 2.22 |
ENSDART00000087724
|
wu:fj05g07
|
wu:fj05g07 |
| chr21_-_39058490 | 2.21 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr8_+_8166285 | 2.20 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr5_+_6617401 | 2.19 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr10_+_16501699 | 2.19 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr6_-_39006449 | 2.18 |
ENSDART00000150885
|
vdrb
|
vitamin D receptor b |
| chr18_-_6803424 | 2.17 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr20_-_51917771 | 2.14 |
ENSDART00000147240
|
dusp10
|
dual specificity phosphatase 10 |
| chr7_-_12596727 | 2.14 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr10_+_41765616 | 2.14 |
ENSDART00000170682
|
rnf34b
|
ring finger protein 34b |
| chr5_-_38506981 | 2.14 |
ENSDART00000097822
|
atp1b2b
|
ATPase Na+/K+ transporting subunit beta 2b |
| chr5_-_33274943 | 2.13 |
ENSDART00000143435
ENSDART00000145222 ENSDART00000004797 |
kyat1
|
kynurenine aminotransferase 1 |
| chr23_-_452365 | 2.13 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr10_-_32524035 | 2.12 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr7_-_38698583 | 2.12 |
ENSDART00000173900
ENSDART00000126737 |
cd59
|
CD59 molecule (CD59 blood group) |
| chr8_+_51958968 | 2.12 |
ENSDART00000139509
ENSDART00000186544 |
gna14a
|
guanine nucleotide binding protein (G protein), alpha 14a |
| chr9_-_10145795 | 2.11 |
ENSDART00000004745
ENSDART00000143295 |
hnmt
|
histamine N-methyltransferase |
| chr2_+_52232630 | 2.10 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr17_+_51764310 | 2.09 |
ENSDART00000157171
|
si:ch211-168d23.3
|
si:ch211-168d23.3 |
| chr19_+_233143 | 2.08 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr14_-_962171 | 2.08 |
ENSDART00000010773
|
acsl1b
|
acyl-CoA synthetase long chain family member 1b |
| chr22_-_38459316 | 2.08 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr2_+_24177006 | 2.07 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr17_+_389218 | 2.07 |
ENSDART00000162898
|
si:rp71-62i8.1
|
si:rp71-62i8.1 |
| chr9_-_48397702 | 2.06 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr6_-_58828398 | 2.05 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr9_-_7655243 | 2.05 |
ENSDART00000102706
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr19_+_342094 | 2.02 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr1_-_12109216 | 2.00 |
ENSDART00000079930
|
mttp
|
microsomal triglyceride transfer protein |
| chr7_+_1579510 | 2.00 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr11_-_38083397 | 1.97 |
ENSDART00000086516
ENSDART00000184033 |
klhdc8a
|
kelch domain containing 8A |
| chr18_-_47662696 | 1.96 |
ENSDART00000184260
|
CABZ01073963.1
|
|
| chr21_+_43253538 | 1.94 |
ENSDART00000179940
ENSDART00000164806 ENSDART00000147026 |
shroom1
|
shroom family member 1 |
| chr10_+_32683089 | 1.94 |
ENSDART00000063551
|
ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr9_-_54840124 | 1.92 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr19_+_1097393 | 1.91 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr22_-_16275236 | 1.90 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr22_-_27136451 | 1.90 |
ENSDART00000153589
|
si:dkey-16m19.1
|
si:dkey-16m19.1 |
| chr13_-_45795722 | 1.89 |
ENSDART00000141779
|
fndc5a
|
fibronectin type III domain containing 5a |
| chr1_-_55196103 | 1.89 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr15_-_9593532 | 1.87 |
ENSDART00000169912
|
gab2
|
GRB2-associated binding protein 2 |
| chr21_-_40880317 | 1.86 |
ENSDART00000100054
ENSDART00000137696 |
elnb
|
elastin b |
| chr10_+_45148005 | 1.85 |
ENSDART00000182501
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr10_-_32524771 | 1.84 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr3_+_472158 | 1.84 |
ENSDART00000134971
|
zgc:194659
|
zgc:194659 |
| chr9_+_993477 | 1.84 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr8_+_8532407 | 1.84 |
ENSDART00000169276
ENSDART00000138993 |
grm6a
|
glutamate receptor, metabotropic 6a |
| chr15_+_20543770 | 1.84 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr25_+_19149241 | 1.84 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr13_+_33688474 | 1.83 |
ENSDART00000161465
|
CABZ01087953.1
|
|
| chr2_-_48826707 | 1.82 |
ENSDART00000134711
|
svilb
|
supervillin b |
| chr22_+_883678 | 1.82 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr7_-_2163361 | 1.81 |
ENSDART00000173654
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr14_-_2933185 | 1.80 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr20_-_53366137 | 1.80 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr19_+_43119698 | 1.79 |
ENSDART00000167847
ENSDART00000186962 ENSDART00000187305 |
eef1a1l1
|
eukaryotic translation elongation factor 1 alpha 1, like 1 |
| chr21_-_32436679 | 1.79 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr19_-_46088429 | 1.76 |
ENSDART00000161385
|
ptdss1b
|
phosphatidylserine synthase 1b |
| chr10_+_45148167 | 1.76 |
ENSDART00000172621
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr1_+_2222363 | 1.76 |
ENSDART00000058877
|
rap2ab
|
RAP2A, member of RAS oncogene family b |
| chr24_-_36238054 | 1.75 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr24_+_3307857 | 1.75 |
ENSDART00000106527
|
gyg1b
|
glycogenin 1b |
| chr18_+_3634652 | 1.73 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr3_+_26019426 | 1.72 |
ENSDART00000135389
ENSDART00000182411 |
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr24_-_20599781 | 1.72 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr3_+_31925067 | 1.72 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr22_-_26558166 | 1.71 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr10_+_41765944 | 1.70 |
ENSDART00000171484
|
rnf34b
|
ring finger protein 34b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.7 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.7 | 5.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 1.3 | 7.5 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 1.2 | 4.9 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 1.1 | 3.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 1.0 | 6.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 1.0 | 4.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 1.0 | 1.9 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.9 | 2.7 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.9 | 2.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.9 | 3.4 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.8 | 3.3 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.7 | 8.1 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.7 | 2.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.7 | 2.7 | GO:0010898 | regulation of triglyceride catabolic process(GO:0010896) positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.6 | 3.1 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.6 | 4.2 | GO:0036268 | swimming(GO:0036268) |
| 0.5 | 3.8 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.5 | 1.5 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.5 | 4.6 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.5 | 9.1 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.5 | 2.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.5 | 2.8 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.5 | 7.0 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.4 | 1.3 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.4 | 1.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 1.7 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.4 | 3.3 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.4 | 4.0 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.4 | 2.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.4 | 3.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.4 | 7.8 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.4 | 2.5 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.4 | 1.8 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.3 | 2.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 1.4 | GO:0070317 | G0 to G1 transition(GO:0045023) regulation of G0 to G1 transition(GO:0070316) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.3 | 4.9 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.3 | 1.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.3 | 2.3 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.3 | 0.9 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.3 | 2.5 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.3 | 1.6 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 1.8 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.3 | 2.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.3 | 6.4 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.3 | 4.5 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 3.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.3 | 4.7 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.3 | 0.9 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.3 | 0.9 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.3 | 0.9 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.3 | 4.6 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 0.8 | GO:0039531 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.3 | 1.7 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.3 | 1.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 2.4 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.3 | 1.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.3 | 2.1 | GO:2000095 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.3 | 1.0 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.2 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.2 | 3.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 2.6 | GO:1902108 | regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902108) |
| 0.2 | 0.7 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 1.2 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.2 | 1.6 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 0.9 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.2 | 1.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.2 | 4.6 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.2 | 3.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.2 | 1.7 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 0.6 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.2 | 1.6 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.2 | 0.6 | GO:0045830 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.2 | 0.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.2 | 1.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 3.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 1.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.2 | 1.6 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.2 | 1.4 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 0.6 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 1.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.2 | 0.9 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.2 | 2.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 2.8 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.2 | 2.2 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.2 | 1.2 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 0.9 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.2 | 0.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 1.6 | GO:0044206 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 0.2 | 0.2 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.2 | 1.3 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 1.3 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 0.8 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.2 | 1.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 3.6 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.2 | 0.5 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 1.2 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.2 | 0.9 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 0.9 | GO:0099525 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.2 | 0.6 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.2 | 0.9 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 3.3 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.2 | 1.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.7 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 1.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 3.0 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 0.4 | GO:0035992 | tendon formation(GO:0035992) |
| 0.1 | 1.1 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 1.2 | GO:0045923 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) positive regulation of fatty acid metabolic process(GO:0045923) |
| 0.1 | 1.3 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 1.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.7 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 1.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.1 | 2.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.1 | 0.4 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.1 | 0.5 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 2.0 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.1 | 0.5 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.1 | 4.9 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.5 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.8 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.1 | 6.5 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.1 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.5 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 0.3 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) semicircular canal fusion(GO:0060879) |
| 0.1 | 3.8 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.1 | 0.4 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.6 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.1 | 1.0 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.7 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.2 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 3.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 2.2 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 0.2 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 1.6 | GO:0006000 | fructose metabolic process(GO:0006000) fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 0.5 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.1 | 0.7 | GO:1901021 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 1.5 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 1.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.2 | GO:0045601 | positive regulation of epithelial cell differentiation(GO:0030858) regulation of endothelial cell differentiation(GO:0045601) |
| 0.1 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.0 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 11.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 2.0 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 2.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.5 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.1 | 0.3 | GO:0048909 | anterior lateral line nerve development(GO:0048909) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.1 | 0.4 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 2.8 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 3.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.1 | 1.3 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.1 | 0.2 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.9 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.3 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.6 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.8 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.1 | 2.9 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.1 | 1.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.8 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.1 | 2.0 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.1 | 0.7 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.1 | 0.3 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 1.0 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 0.3 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 1.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.4 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:1903644 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.1 | 0.4 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.1 | 1.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 0.3 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.3 | GO:0061551 | trigeminal ganglion development(GO:0061551) |
| 0.1 | 5.4 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.1 | 0.5 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 0.5 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 3.0 | GO:0035304 | regulation of protein dephosphorylation(GO:0035304) |
| 0.1 | 0.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.4 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.5 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 2.9 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 2.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.1 | 0.8 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.1 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.2 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 1.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.1 | 0.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.1 | 1.0 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.4 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.1 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.2 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 1.8 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 0.4 | GO:0050890 | cognition(GO:0050890) |
| 0.1 | 2.5 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 | 1.9 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.1 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.8 | GO:0097581 | lamellipodium organization(GO:0097581) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.0 | 0.2 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.0 | 0.6 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.3 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.9 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 5.2 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.9 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.4 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.5 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.2 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.0 | 2.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.4 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.5 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.4 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 2.1 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.0 | 0.9 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.2 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 3.8 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.5 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.9 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 2.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.3 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.4 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 2.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.6 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.5 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.5 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.9 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 1.7 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.9 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.6 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 3.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:2000812 | positive regulation of protein depolymerization(GO:1901881) regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.6 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.3 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.0 | 0.8 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.4 | GO:0006112 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) energy reserve metabolic process(GO:0006112) glucan metabolic process(GO:0044042) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 2.0 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 0.9 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 1.2 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.7 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.8 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 3.3 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 1.6 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.2 | GO:0046473 | phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.3 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.9 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.0 | 0.1 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.6 | GO:0003140 | determination of left/right asymmetry in lateral mesoderm(GO:0003140) |
| 0.0 | 0.8 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 3.6 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.3 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.4 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 1.3 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 1.0 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.5 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.1 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0006559 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.5 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.4 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0019859 | thymine catabolic process(GO:0006210) valine catabolic process(GO:0006574) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.2 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) |
| 0.0 | 0.6 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.2 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0009154 | purine ribonucleotide catabolic process(GO:0009154) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 15.5 | GO:0070062 | extracellular exosome(GO:0070062) |
| 1.4 | 6.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.8 | 3.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 3.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.4 | 5.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.3 | 3.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.3 | 1.9 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.3 | 3.3 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.3 | 1.7 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.3 | 2.9 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.3 | 4.1 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 4.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.2 | 1.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 2.7 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.2 | 2.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 0.6 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.2 | 0.6 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 3.8 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 8.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 0.5 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.2 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 1.0 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 3.0 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.4 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 2.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.8 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 4.0 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.1 | 1.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 3.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 5.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 5.0 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 2.9 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 5.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.7 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.0 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.3 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 0.6 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 10.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 6.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 1.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 6.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.9 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 0.9 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.0 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 1.0 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 2.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 1.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 2.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 4.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 14.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.4 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 2.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.9 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 4.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 2.0 | GO:0034703 | cation channel complex(GO:0034703) |
| 0.0 | 1.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 2.6 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.1 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.5 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 1.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.8 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 1.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.3 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.4 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.9 | GO:0030117 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.4 | 4.2 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 1.2 | 4.8 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 1.1 | 3.3 | GO:0034618 | arginine binding(GO:0034618) |
| 1.1 | 7.5 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 1.0 | 5.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.0 | 7.6 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.9 | 4.5 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.7 | 2.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.6 | 2.4 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.6 | 2.4 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.6 | 5.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.6 | 7.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.6 | 3.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.6 | 3.3 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.5 | 3.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.5 | 2.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.5 | 4.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 2.0 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.5 | 2.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.5 | 4.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.5 | 3.4 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.5 | 3.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 1.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.4 | 1.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.4 | 0.4 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 4.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.4 | 3.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.4 | 2.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.4 | 5.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.4 | 1.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.4 | 10.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 4.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.4 | 1.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.4 | 2.8 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 1.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 2.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.8 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.3 | 2.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 16.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.3 | 0.9 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.3 | 3.7 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.3 | 1.8 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.3 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 1.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 3.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.2 | 1.0 | GO:0031781 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.2 | 2.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 3.0 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.2 | 0.7 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.2 | 2.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 2.2 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 1.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.2 | 2.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.2 | 1.1 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 6.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 3.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.6 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.2 | 1.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 2.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 1.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 0.6 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 2.1 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 3.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.2 | 1.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 0.5 | GO:0031779 | melanocortin receptor binding(GO:0031779) |
| 0.2 | 0.9 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.2 | 1.3 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 4.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.2 | 1.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 0.5 | GO:1990174 | phosphodiesterase decapping endonuclease activity(GO:1990174) |
| 0.2 | 3.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.2 | 0.5 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.2 | 1.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 6.0 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 1.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 1.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 4.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 3.9 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 3.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 4.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.5 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.5 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 1.9 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.1 | 2.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.6 | GO:0008443 | phosphofructokinase activity(GO:0008443) |
| 0.1 | 0.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 0.4 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 1.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 1.0 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.9 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.5 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.9 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 2.6 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.1 | 0.3 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.4 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.4 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.3 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.1 | 0.9 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 2.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 1.0 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 2.0 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 1.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 7.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.3 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.1 | 1.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 2.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.1 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.6 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.7 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.4 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.1 | 0.9 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.7 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 4.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.8 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 4.5 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 1.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.9 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.5 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 1.0 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 4.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 2.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 15.1 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.2 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.0 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 2.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.4 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.2 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 1.0 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 1.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.4 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.2 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.7 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.1 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0072571 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.0 | 0.5 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.1 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 1.8 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 8.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.3 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 2.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 1.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.6 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.2 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.3 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.3 | 3.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.2 | 1.6 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 2.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.2 | 3.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 0.8 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.2 | 3.2 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 0.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 1.9 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.1 | 1.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 1.7 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 1.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 1.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 1.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.5 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.1 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.2 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 3.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 1.4 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.1 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 2.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 2.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 4.3 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.3 | 2.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 7.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.3 | 2.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 2.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 1.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 3.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 2.0 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.2 | 2.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 1.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.2 | 2.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.2 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 5.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.1 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 2.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 2.4 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 4.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.9 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.1 | 1.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 2.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.5 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.9 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 1.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 3.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 1.7 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 0.8 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 3.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.0 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.1 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 1.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PI3K EVENTS IN ERBB4 SIGNALING | Genes involved in PI3K events in ERBB4 signaling |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.4 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.5 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.2 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.7 | REACTOME ION CHANNEL TRANSPORT | Genes involved in Ion channel transport |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.5 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.9 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |