Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
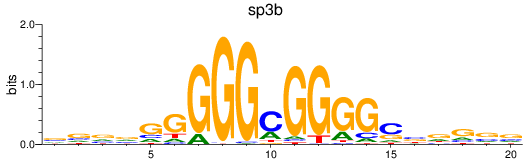
Results for sp3b
Z-value: 0.94
Transcription factors associated with sp3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp3b
|
ENSDARG00000007812 | Sp3b transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp3b | dr11_v1_chr6_-_10740365_10740365 | 0.71 | 6.4e-04 | Click! |
Activity profile of sp3b motif
Sorted Z-values of sp3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77557279 | 3.98 |
ENSDART00000180113
|
AL935186.10
|
|
| chr5_-_1047222 | 2.49 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr13_+_1100197 | 2.05 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr3_-_62380146 | 1.95 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr2_+_58221163 | 1.95 |
ENSDART00000157939
|
FO704813.1
|
|
| chr8_+_43056153 | 1.80 |
ENSDART00000186377
|
prnpa
|
prion protein a |
| chr14_-_2933185 | 1.54 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr17_+_12075805 | 1.47 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr8_+_1082100 | 1.46 |
ENSDART00000149276
|
lzts3b
|
leucine zipper, putative tumor suppressor family member 3b |
| chr6_-_60104628 | 1.31 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr3_-_16227683 | 1.28 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr24_-_14711597 | 1.24 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr20_+_46741074 | 1.23 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr19_+_935565 | 1.20 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr25_+_16356083 | 1.20 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr4_+_3358383 | 1.18 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr8_+_2656231 | 1.15 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr14_+_30910114 | 1.13 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr21_+_1143141 | 1.12 |
ENSDART00000178294
|
CABZ01086139.1
|
|
| chr10_-_641609 | 1.12 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr7_+_74141297 | 1.08 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr3_+_49043917 | 1.05 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr13_-_17860307 | 1.04 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr16_-_8120203 | 1.02 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr23_-_30960506 | 1.01 |
ENSDART00000142661
|
osbpl2a
|
oxysterol binding protein-like 2a |
| chr25_-_35599887 | 1.01 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr21_-_39058490 | 1.01 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr13_+_3819475 | 1.01 |
ENSDART00000139958
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr7_-_6592142 | 1.01 |
ENSDART00000160137
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr13_-_638485 | 1.01 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr8_+_6576940 | 1.00 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr6_-_58828398 | 0.97 |
ENSDART00000090634
|
kif5ab
|
kinesin family member 5A, b |
| chr19_+_156757 | 0.96 |
ENSDART00000167717
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr5_+_6617401 | 0.96 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr23_+_17220986 | 0.95 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr20_+_13175379 | 0.94 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr15_-_47956388 | 0.93 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr7_-_61683417 | 0.93 |
ENSDART00000098622
ENSDART00000184088 ENSDART00000148270 |
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr10_+_44924684 | 0.93 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
| chr10_+_5135842 | 0.92 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr9_-_54840124 | 0.92 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr2_-_44282796 | 0.92 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr4_-_68831753 | 0.91 |
ENSDART00000189170
|
si:dkey-264f17.1
|
si:dkey-264f17.1 |
| chr2_+_24177190 | 0.91 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr20_+_52389858 | 0.90 |
ENSDART00000185863
ENSDART00000166651 |
arhgap39
|
Rho GTPase activating protein 39 |
| chr4_-_22671469 | 0.89 |
ENSDART00000050753
|
cd36
|
CD36 molecule (thrombospondin receptor) |
| chr2_+_48073972 | 0.89 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr2_+_258698 | 0.89 |
ENSDART00000181330
ENSDART00000181645 |
phlpp1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr16_+_34528409 | 0.88 |
ENSDART00000144718
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr7_+_7630409 | 0.87 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr6_-_38818582 | 0.86 |
ENSDART00000149833
|
cnga3a
|
cyclic nucleotide gated channel alpha 3a |
| chr3_-_58455289 | 0.86 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr16_+_4838808 | 0.85 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr8_-_53490376 | 0.84 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr5_-_13086616 | 0.84 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr4_+_55758103 | 0.83 |
ENSDART00000185964
|
CT583728.23
|
|
| chr5_+_51443009 | 0.83 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr12_-_45349849 | 0.82 |
ENSDART00000183036
|
CABZ01068367.1
|
Danio rerio uncharacterized LOC100332446 (LOC100332446), mRNA. |
| chr25_-_30047477 | 0.81 |
ENSDART00000164859
|
CR759810.1
|
|
| chr3_+_11548516 | 0.81 |
ENSDART00000059117
|
mmd
|
monocyte to macrophage differentiation-associated |
| chr23_+_45611649 | 0.80 |
ENSDART00000169521
|
dclk2b
|
doublecortin-like kinase 2b |
| chr6_+_2271559 | 0.80 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr6_+_34511886 | 0.78 |
ENSDART00000179450
|
lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr20_+_48413712 | 0.78 |
ENSDART00000159983
|
CABZ01059119.1
|
|
| chr7_+_67325933 | 0.78 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr8_+_8166285 | 0.77 |
ENSDART00000147940
|
plxnb3
|
plexin B3 |
| chr3_+_12322170 | 0.77 |
ENSDART00000161227
|
glis2b
|
GLIS family zinc finger 2b |
| chr24_-_26622423 | 0.77 |
ENSDART00000182044
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr15_-_44601331 | 0.76 |
ENSDART00000161514
|
zgc:165508
|
zgc:165508 |
| chr15_-_43768776 | 0.76 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr6_-_1874664 | 0.76 |
ENSDART00000007972
|
dlgap4b
|
discs, large (Drosophila) homolog-associated protein 4b |
| chr8_+_22485956 | 0.75 |
ENSDART00000099960
|
si:ch211-261n11.7
|
si:ch211-261n11.7 |
| chr10_+_12517059 | 0.75 |
ENSDART00000081140
|
CT990561.1
|
|
| chr23_-_900795 | 0.75 |
ENSDART00000190517
ENSDART00000182849 ENSDART00000111456 ENSDART00000185430 |
rbm10
|
RNA binding motif protein 10 |
| chr10_+_33895315 | 0.73 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr9_-_33725972 | 0.73 |
ENSDART00000028225
|
mao
|
monoamine oxidase |
| chr12_+_49125510 | 0.72 |
ENSDART00000185804
|
FO704607.1
|
|
| chr9_+_993477 | 0.72 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr2_-_4874966 | 0.72 |
ENSDART00000165296
|
tfr1a
|
transferrin receptor 1a |
| chr25_-_19395476 | 0.71 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr2_-_58075414 | 0.71 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr1_+_54677173 | 0.71 |
ENSDART00000114705
|
gprc5bb
|
G protein-coupled receptor, class C, group 5, member Bb |
| chr21_-_45891262 | 0.71 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr5_-_69041102 | 0.70 |
ENSDART00000161561
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr1_+_9004719 | 0.69 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr4_-_23839789 | 0.68 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr9_+_36946340 | 0.68 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr3_+_32526799 | 0.68 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr16_+_4839078 | 0.67 |
ENSDART00000150111
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr16_-_53919115 | 0.67 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| chr5_+_72152813 | 0.66 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr7_+_1579510 | 0.66 |
ENSDART00000190525
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr12_+_5530247 | 0.66 |
ENSDART00000114637
|
ace
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| chr6_-_8580857 | 0.65 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr15_-_37543591 | 0.65 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr23_+_14217508 | 0.65 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr8_+_1187928 | 0.64 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr15_+_47582207 | 0.64 |
ENSDART00000159388
|
CABZ01087566.1
|
|
| chr1_-_31505144 | 0.63 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr1_-_57501299 | 0.63 |
ENSDART00000080600
|
zgc:171470
|
zgc:171470 |
| chr3_+_15550522 | 0.62 |
ENSDART00000136912
ENSDART00000176218 |
si:dkey-93n13.3
|
si:dkey-93n13.3 |
| chr18_+_26086803 | 0.62 |
ENSDART00000187911
|
znf710a
|
zinc finger protein 710a |
| chr5_-_19861766 | 0.62 |
ENSDART00000088904
|
gltpa
|
glycolipid transfer protein a |
| chr6_-_58828113 | 0.61 |
ENSDART00000180934
|
kif5ab
|
kinesin family member 5A, b |
| chr1_+_6307305 | 0.61 |
ENSDART00000183870
|
LO018629.1
|
|
| chr18_-_8030073 | 0.60 |
ENSDART00000151490
|
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr13_-_5978433 | 0.60 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr17_-_22048233 | 0.60 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr15_-_33925851 | 0.59 |
ENSDART00000187807
ENSDART00000187780 |
mag
|
myelin associated glycoprotein |
| chr15_-_2726662 | 0.59 |
ENSDART00000108856
|
si:ch211-235m3.5
|
si:ch211-235m3.5 |
| chr11_+_36989696 | 0.59 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr13_-_27767330 | 0.59 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr10_+_1638876 | 0.59 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr21_-_32436679 | 0.58 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr13_-_51922290 | 0.57 |
ENSDART00000168648
|
srfb
|
serum response factor b |
| chr4_-_68569527 | 0.56 |
ENSDART00000192091
|
BX548011.5
|
|
| chr5_-_71838520 | 0.56 |
ENSDART00000174396
|
CU927890.1
|
|
| chr8_-_410728 | 0.55 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr20_-_16972351 | 0.55 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr6_+_102506 | 0.55 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr22_-_7563593 | 0.54 |
ENSDART00000106062
|
BX511034.1
|
|
| chr3_+_59851537 | 0.54 |
ENSDART00000180997
|
CU693479.1
|
|
| chr7_-_55454406 | 0.54 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr13_-_21739142 | 0.54 |
ENSDART00000078460
|
si:dkey-191g9.5
|
si:dkey-191g9.5 |
| chr19_+_342094 | 0.53 |
ENSDART00000151013
ENSDART00000187622 |
ensaa
|
endosulfine alpha a |
| chr16_-_280835 | 0.53 |
ENSDART00000190541
|
LO017917.1
|
|
| chr25_+_4581214 | 0.53 |
ENSDART00000185552
|
CABZ01068600.1
|
|
| chr25_-_36492779 | 0.52 |
ENSDART00000042271
|
irx3b
|
iroquois homeobox 3b |
| chr8_+_28900689 | 0.52 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr22_+_883678 | 0.52 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr25_+_336503 | 0.52 |
ENSDART00000160395
|
CU929262.1
|
|
| chr19_-_6193448 | 0.52 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr18_-_46763170 | 0.52 |
ENSDART00000171880
|
dner
|
delta/notch-like EGF repeat containing |
| chr11_+_45287541 | 0.51 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr19_-_44091405 | 0.51 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr20_-_33675676 | 0.51 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr17_+_25856671 | 0.50 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr25_+_34407529 | 0.50 |
ENSDART00000156751
|
OTUD7A
|
si:dkey-37f18.2 |
| chr5_-_14326959 | 0.50 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr22_-_34979139 | 0.50 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr8_-_22739757 | 0.50 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr7_-_23768234 | 0.50 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr15_+_20543770 | 0.50 |
ENSDART00000092357
|
sgsm2
|
small G protein signaling modulator 2 |
| chr3_-_18274691 | 0.50 |
ENSDART00000161140
|
BX649434.3
|
|
| chr21_-_45086170 | 0.50 |
ENSDART00000188963
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr3_+_40841897 | 0.50 |
ENSDART00000150081
ENSDART00000126096 |
mmd2a
|
monocyte to macrophage differentiation-associated 2a |
| chr8_+_48965767 | 0.49 |
ENSDART00000008058
|
aak1a
|
AP2 associated kinase 1a |
| chr25_+_37443194 | 0.49 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr20_+_39250673 | 0.49 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr22_-_607812 | 0.48 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr10_+_44719697 | 0.48 |
ENSDART00000158087
|
scarb1
|
scavenger receptor class B, member 1 |
| chr19_-_22498629 | 0.48 |
ENSDART00000185370
|
pleca
|
plectin a |
| chr18_-_14941840 | 0.48 |
ENSDART00000091729
|
mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr25_+_35706493 | 0.47 |
ENSDART00000176741
|
kif21a
|
kinesin family member 21A |
| chr18_+_660578 | 0.47 |
ENSDART00000161203
|
CELF6
|
si:dkey-205h23.2 |
| chr15_-_17025212 | 0.47 |
ENSDART00000014465
|
hip1
|
huntingtin interacting protein 1 |
| chr9_+_30274637 | 0.46 |
ENSDART00000079095
|
rpgra
|
retinitis pigmentosa GTPase regulator a |
| chr18_+_507835 | 0.46 |
ENSDART00000189701
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr3_-_5964557 | 0.46 |
ENSDART00000184738
|
BX284638.1
|
|
| chr9_-_2572790 | 0.46 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr6_-_21830405 | 0.46 |
ENSDART00000151803
ENSDART00000113497 |
setd5
|
SET domain containing 5 |
| chr7_-_5148388 | 0.46 |
ENSDART00000143549
|
si:ch211-272h9.5
|
si:ch211-272h9.5 |
| chr17_-_2721336 | 0.46 |
ENSDART00000109582
ENSDART00000192691 ENSDART00000189381 |
spata7
|
spermatogenesis associated 7 |
| chr10_+_10210455 | 0.46 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr8_+_29963024 | 0.46 |
ENSDART00000149372
|
ptch1
|
patched 1 |
| chr21_+_28445052 | 0.45 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr15_+_45544589 | 0.45 |
ENSDART00000055978
|
LO018197.1
|
|
| chr9_+_55154414 | 0.45 |
ENSDART00000182924
|
ANOS1
|
anosmin 1 |
| chr23_-_1660708 | 0.45 |
ENSDART00000175138
|
CU693481.1
|
|
| chr6_+_48206535 | 0.45 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr17_+_44756247 | 0.44 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr4_-_37766174 | 0.44 |
ENSDART00000170066
|
si:dkey-207l24.2
|
si:dkey-207l24.2 |
| chr3_+_22079219 | 0.44 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr20_-_22378401 | 0.44 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr8_-_5847533 | 0.43 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr22_-_18112374 | 0.43 |
ENSDART00000191154
|
ncanb
|
neurocan b |
| chr8_+_51958968 | 0.43 |
ENSDART00000139509
ENSDART00000186544 |
gna14a
|
guanine nucleotide binding protein (G protein), alpha 14a |
| chr2_+_43469241 | 0.43 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr2_+_24177006 | 0.43 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr8_-_53488832 | 0.43 |
ENSDART00000191801
|
chdh
|
choline dehydrogenase |
| chr14_+_15620640 | 0.43 |
ENSDART00000188867
|
si:dkey-203a12.9
|
si:dkey-203a12.9 |
| chr8_+_7740004 | 0.43 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr1_+_24469313 | 0.42 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr5_-_5243079 | 0.42 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr24_+_40473032 | 0.42 |
ENSDART00000084238
ENSDART00000178508 |
CABZ01076968.1
|
|
| chr5_-_30074332 | 0.41 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr9_+_15890558 | 0.41 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr15_-_14552101 | 0.41 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr4_+_3980247 | 0.41 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr6_+_48862 | 0.41 |
ENSDART00000082954
|
mbd5
|
methyl-CpG binding domain protein 5 |
| chr9_-_11263228 | 0.41 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr16_-_8613499 | 0.41 |
ENSDART00000189189
|
cobl
|
cordon-bleu WH2 repeat protein |
| chr25_-_10724947 | 0.40 |
ENSDART00000184490
|
lrp5
|
low density lipoprotein receptor-related protein 5 |
| chr16_-_52733384 | 0.40 |
ENSDART00000147236
ENSDART00000056101 |
azin1a
|
antizyme inhibitor 1a |
| chr11_-_44030962 | 0.40 |
ENSDART00000171910
|
FP016005.1
|
|
| chr14_-_448182 | 0.40 |
ENSDART00000180018
|
FAT4
|
FAT atypical cadherin 4 |
| chr8_+_48966165 | 0.40 |
ENSDART00000165425
|
aak1a
|
AP2 associated kinase 1a |
| chr6_+_58915889 | 0.40 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr16_-_20707742 | 0.39 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr4_+_5537101 | 0.39 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.4 | 1.3 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.4 | 1.1 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) |
| 0.3 | 1.0 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 0.3 | GO:1905067 | negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.3 | 1.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.3 | 0.8 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 1.6 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 1.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.2 | 0.7 | GO:0015682 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.2 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 0.5 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.2 | 0.5 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.2 | 0.5 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.2 | 0.5 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.1 | 0.6 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.1 | 1.0 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 0.3 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.1 | 0.4 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 0.8 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 0.9 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.1 | 0.4 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.8 | GO:2000463 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 0.7 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.4 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.4 | GO:1902590 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.5 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.1 | 0.4 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.1 | 1.5 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.1 | 0.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.0 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.1 | 0.8 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.1 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 0.7 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.7 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.1 | 0.3 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.6 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 1.6 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.1 | 0.5 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 0.9 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.5 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 1.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.1 | 0.5 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 1.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.2 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.1 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 0.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 0.9 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.8 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.4 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.5 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.3 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.0 | 1.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 1.6 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 1.0 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.9 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.8 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 1.0 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0061384 | trabecula morphogenesis(GO:0061383) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.8 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.4 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.4 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 1.0 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.2 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.7 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.2 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.5 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.5 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.0 | 0.1 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.0 | 0.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 1.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 1.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.4 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.5 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 3.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.5 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.4 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.4 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.5 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.7 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 1.0 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.7 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.7 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.8 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.8 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 1.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 1.8 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 0.8 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.2 | 0.7 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.2 | 0.6 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.7 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.8 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.0 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.3 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.5 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.4 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.7 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.7 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.9 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.9 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.1 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 1.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.4 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 1.0 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 1.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 2.1 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 1.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.3 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 1.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.4 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 1.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0030955 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.6 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.5 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.8 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 2.0 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 1.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 1.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.9 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 0.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.1 | 1.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 1.6 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.9 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |