Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
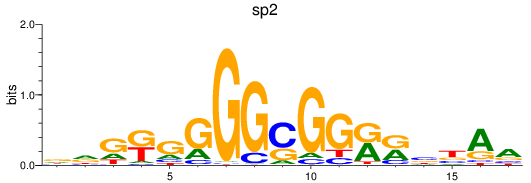
Results for sp2
Z-value: 1.60
Transcription factors associated with sp2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp2
|
ENSDARG00000076763 | sp2 transcription factor |
|
sp2
|
ENSDARG00000113443 | sp2 transcription factor |
|
sp2
|
ENSDARG00000115477 | sp2 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp2 | dr11_v1_chr11_-_12008001_12008001 | -0.05 | 8.3e-01 | Click! |
Activity profile of sp2 motif
Sorted Z-values of sp2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_16941319 | 8.67 |
ENSDART00000109968
|
zgc:174855
|
zgc:174855 |
| chr23_-_46201008 | 8.32 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr12_-_16764751 | 7.76 |
ENSDART00000113862
|
zgc:174154
|
zgc:174154 |
| chr18_-_46010 | 7.38 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr9_+_15890558 | 5.63 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr16_-_7793457 | 4.87 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr20_+_33987465 | 4.73 |
ENSDART00000061751
|
zp3a.2
|
zona pellucida glycoprotein 3a, tandem duplicate 2 |
| chr18_-_127558 | 4.64 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr5_+_43870389 | 4.44 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr5_+_23136544 | 4.16 |
ENSDART00000003428
ENSDART00000109340 ENSDART00000171039 ENSDART00000178821 |
prps1a
|
phosphoribosyl pyrophosphate synthetase 1A |
| chr10_+_10788811 | 4.05 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr18_-_127873 | 4.03 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr14_-_12822 | 3.89 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr16_+_46294337 | 3.83 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr6_+_12922002 | 3.45 |
ENSDART00000080350
ENSDART00000149018 |
cxcr4a
|
chemokine (C-X-C motif) receptor 4a |
| chr17_+_51746830 | 3.19 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr20_+_33991801 | 3.18 |
ENSDART00000061744
|
zp3a.1
|
zona pellucida glycoprotein 3a, tandem duplicate 1 |
| chr3_-_32541033 | 3.14 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr25_+_22319940 | 3.05 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr7_-_32020100 | 3.00 |
ENSDART00000185433
|
kif18a
|
kinesin family member 18A |
| chr20_+_54304800 | 2.90 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr5_-_14326959 | 2.77 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr1_-_59232267 | 2.76 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr20_+_18740518 | 2.68 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr13_-_12021566 | 2.67 |
ENSDART00000125430
|
pprc1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1 |
| chr1_-_45616470 | 2.67 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_51157660 | 2.61 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr19_-_15855427 | 2.57 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr8_+_8936912 | 2.50 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr17_-_2584423 | 2.44 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_+_35387517 | 2.43 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr8_+_39802506 | 2.42 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr7_-_16598212 | 2.40 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr23_+_31107685 | 2.38 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr19_+_6990970 | 2.37 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr18_-_50799510 | 2.33 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr14_+_31865324 | 2.32 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr22_-_5682494 | 2.31 |
ENSDART00000012686
|
dnase1l4.1
|
deoxyribonuclease 1 like 4, tandem duplicate 1 |
| chr21_-_43117327 | 2.31 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr17_-_48944465 | 2.31 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr17_-_37474689 | 2.28 |
ENSDART00000103980
|
crip2
|
cysteine-rich protein 2 |
| chr22_+_465269 | 2.28 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr2_+_58841181 | 2.26 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr18_-_45736 | 2.26 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr21_-_22325124 | 2.23 |
ENSDART00000142100
|
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr23_-_2513300 | 2.19 |
ENSDART00000067652
|
snai1b
|
snail family zinc finger 1b |
| chr1_-_45616242 | 2.13 |
ENSDART00000150066
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr15_-_47865063 | 2.13 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr5_+_36899691 | 2.13 |
ENSDART00000132322
|
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr7_+_41314862 | 2.10 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr7_-_7493758 | 2.07 |
ENSDART00000036703
|
pfdn2
|
prefoldin subunit 2 |
| chr11_-_16395956 | 2.04 |
ENSDART00000115085
|
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr7_+_22792132 | 2.02 |
ENSDART00000135207
ENSDART00000146801 |
rbm4.3
|
RNA binding motif protein 4.3 |
| chr8_-_2591654 | 2.01 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr14_+_28545198 | 1.98 |
ENSDART00000125362
ENSDART00000105902 |
hmmr
|
hyaluronan-mediated motility receptor (RHAMM) |
| chr4_-_5795309 | 1.97 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr2_-_44282796 | 1.96 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr3_-_33941319 | 1.93 |
ENSDART00000026090
ENSDART00000111878 |
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr20_+_6630540 | 1.93 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr3_-_33941875 | 1.92 |
ENSDART00000047660
|
gtf2f1
|
general transcription factor IIF, polypeptide 1 |
| chr5_+_12743640 | 1.89 |
ENSDART00000081411
|
pole
|
polymerase (DNA directed), epsilon |
| chr2_+_23790748 | 1.87 |
ENSDART00000041877
|
csrnp1a
|
cysteine-serine-rich nuclear protein 1a |
| chr20_+_36623807 | 1.86 |
ENSDART00000149171
ENSDART00000062895 |
srp9
|
signal recognition particle 9 |
| chr23_+_30898013 | 1.86 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr13_-_45022301 | 1.86 |
ENSDART00000183589
ENSDART00000125633 ENSDART00000074787 |
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr8_-_53490376 | 1.85 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr14_+_31865099 | 1.84 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr14_-_237130 | 1.84 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr19_+_12406583 | 1.83 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr17_+_27723490 | 1.83 |
ENSDART00000123588
ENSDART00000170462 ENSDART00000169708 |
qkia
|
QKI, KH domain containing, RNA binding a |
| chr24_+_38671054 | 1.81 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr3_+_35812040 | 1.78 |
ENSDART00000075903
ENSDART00000147712 |
crlf3
|
cytokine receptor-like factor 3 |
| chr25_+_1732838 | 1.77 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr7_-_45852270 | 1.76 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr1_-_51157454 | 1.75 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr12_-_42214 | 1.70 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr13_-_45022527 | 1.70 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr1_+_54115839 | 1.69 |
ENSDART00000180214
|
LO017722.2
|
|
| chr10_-_44924289 | 1.69 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr3_-_40276057 | 1.69 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr9_+_23825440 | 1.65 |
ENSDART00000138470
|
ints6
|
integrator complex subunit 6 |
| chr3_+_46559639 | 1.63 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr9_+_23003208 | 1.63 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr5_+_36900157 | 1.63 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr21_+_5589923 | 1.63 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr8_+_2456854 | 1.62 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr7_-_48396193 | 1.61 |
ENSDART00000083555
|
sin3ab
|
SIN3 transcription regulator family member Ab |
| chr20_+_39250673 | 1.60 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr11_-_36341028 | 1.60 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr23_+_44374041 | 1.60 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr18_-_15467446 | 1.56 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr3_+_35611625 | 1.56 |
ENSDART00000190995
|
traf7
|
TNF receptor-associated factor 7 |
| chr7_-_6444011 | 1.55 |
ENSDART00000173010
|
zgc:112234
|
zgc:112234 |
| chr21_+_28747069 | 1.55 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr14_-_42231293 | 1.52 |
ENSDART00000185486
|
BX890543.1
|
|
| chr7_-_13906409 | 1.50 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr15_-_25093680 | 1.48 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr10_-_43113731 | 1.47 |
ENSDART00000138099
|
tmem167a
|
transmembrane protein 167A |
| chr23_+_2825940 | 1.47 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr15_-_20468302 | 1.47 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr2_+_105748 | 1.47 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr8_-_41279326 | 1.46 |
ENSDART00000075491
|
pop5
|
POP5 homolog, ribonuclease P/MRP subunit |
| chr17_+_49081828 | 1.45 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr11_+_7432533 | 1.44 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr7_+_55112922 | 1.39 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr19_-_26769867 | 1.39 |
ENSDART00000043776
ENSDART00000159489 ENSDART00000138675 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr3_+_16265924 | 1.38 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr22_-_6651382 | 1.37 |
ENSDART00000124308
|
si:ch211-209l18.4
|
si:ch211-209l18.4 |
| chr10_-_3332362 | 1.37 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr13_+_16279890 | 1.36 |
ENSDART00000101775
ENSDART00000057948 |
anxa11a
|
annexin A11a |
| chr18_-_3166726 | 1.36 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr12_+_14149686 | 1.36 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr10_-_42297889 | 1.34 |
ENSDART00000099262
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr5_-_22619879 | 1.34 |
ENSDART00000051623
|
zgc:113208
|
zgc:113208 |
| chr25_-_28674739 | 1.34 |
ENSDART00000067073
|
lrrc10
|
leucine rich repeat containing 10 |
| chr3_+_13603272 | 1.32 |
ENSDART00000185084
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr19_-_26770083 | 1.32 |
ENSDART00000193811
ENSDART00000174455 |
prrc2a
|
proline-rich coiled-coil 2A |
| chr1_+_604127 | 1.31 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr25_+_3104959 | 1.30 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr19_+_4912817 | 1.29 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr7_+_38897836 | 1.29 |
ENSDART00000024330
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr2_+_55365727 | 1.29 |
ENSDART00000162943
|
FP245456.1
|
|
| chr21_+_5129513 | 1.29 |
ENSDART00000102572
|
thbs4b
|
thrombospondin 4b |
| chr1_-_12393060 | 1.27 |
ENSDART00000079782
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr3_-_5964557 | 1.26 |
ENSDART00000184738
|
BX284638.1
|
|
| chr21_+_249970 | 1.24 |
ENSDART00000169026
|
jak2a
|
Janus kinase 2a |
| chr15_-_25094026 | 1.24 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr14_-_9522364 | 1.21 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr11_+_19370447 | 1.18 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr18_-_34549721 | 1.17 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr7_-_66126628 | 1.17 |
ENSDART00000184492
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr18_+_46151505 | 1.16 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr2_+_16696052 | 1.15 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr11_-_36341189 | 1.14 |
ENSDART00000159752
|
sort1a
|
sortilin 1a |
| chr24_-_36593876 | 1.12 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr17_-_43287290 | 1.10 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr12_-_17492852 | 1.10 |
ENSDART00000012421
ENSDART00000138766 ENSDART00000130735 |
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr19_+_4892281 | 1.10 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr25_-_6389713 | 1.10 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr1_+_13087797 | 1.09 |
ENSDART00000170314
|
si:dkey-5n7.2
|
si:dkey-5n7.2 |
| chr20_-_16156419 | 1.08 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_+_39200213 | 1.08 |
ENSDART00000153727
|
ei24
|
etoposide induced 2.4 |
| chr18_-_35842554 | 1.07 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr14_-_46113321 | 1.06 |
ENSDART00000169040
ENSDART00000161475 ENSDART00000124925 |
si:ch211-235e9.8
|
si:ch211-235e9.8 |
| chr22_+_24215007 | 1.06 |
ENSDART00000162227
|
glrx2
|
glutaredoxin 2 |
| chr6_+_3334710 | 1.06 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr6_-_23117348 | 1.05 |
ENSDART00000154941
ENSDART00000154252 |
ten1
|
TEN1 CST complex subunit |
| chr14_-_15154695 | 1.04 |
ENSDART00000160677
|
uvssa
|
UV-stimulated scaffold protein A |
| chr5_-_16351306 | 1.04 |
ENSDART00000168643
|
CABZ01088700.1
|
|
| chr14_-_6225336 | 1.03 |
ENSDART00000111681
|
hdx
|
highly divergent homeobox |
| chr9_+_30464641 | 1.02 |
ENSDART00000128357
|
gja5a
|
gap junction protein, alpha 5a |
| chr12_+_39203745 | 1.02 |
ENSDART00000153661
|
si:dkeyp-106c3.2
|
si:dkeyp-106c3.2 |
| chr10_-_641609 | 1.01 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr19_-_2707048 | 1.00 |
ENSDART00000166112
|
il6
|
interleukin 6 (interferon, beta 2) |
| chr15_+_1397811 | 1.00 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr13_-_26799244 | 0.99 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr8_+_27555314 | 0.98 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr25_+_3507368 | 0.98 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr25_+_16356083 | 0.97 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr21_-_43328056 | 0.96 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr19_-_31802296 | 0.96 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr23_+_4890693 | 0.94 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr4_-_11064073 | 0.94 |
ENSDART00000150760
|
si:dkey-21h14.8
|
si:dkey-21h14.8 |
| chr16_-_7227021 | 0.94 |
ENSDART00000187916
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr4_-_34799395 | 0.94 |
ENSDART00000170744
|
si:dkey-146m20.13
|
si:dkey-146m20.13 |
| chr7_+_38898208 | 0.93 |
ENSDART00000172251
|
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr19_+_636886 | 0.92 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr21_+_28747236 | 0.92 |
ENSDART00000137874
|
zgc:100829
|
zgc:100829 |
| chr18_-_26715156 | 0.90 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr15_+_25489406 | 0.89 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr5_-_38170996 | 0.89 |
ENSDART00000145805
|
si:ch211-284e13.12
|
si:ch211-284e13.12 |
| chr23_-_45407631 | 0.87 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr20_+_34868933 | 0.87 |
ENSDART00000153006
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr3_+_16841942 | 0.87 |
ENSDART00000023985
ENSDART00000145317 |
stk17al
|
serine/threonine kinase 17a like |
| chr4_-_72468562 | 0.86 |
ENSDART00000181890
|
CR788316.1
|
|
| chr18_-_22094102 | 0.84 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr8_+_23152244 | 0.83 |
ENSDART00000036801
ENSDART00000184298 |
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr23_-_13875252 | 0.83 |
ENSDART00000104834
ENSDART00000193807 |
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr24_+_42132962 | 0.82 |
ENSDART00000187739
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr25_+_19008497 | 0.82 |
ENSDART00000104420
|
samm50
|
SAMM50 sorting and assembly machinery component |
| chr15_-_6976851 | 0.81 |
ENSDART00000158474
ENSDART00000168943 ENSDART00000169944 |
si:ch73-311h14.2
|
si:ch73-311h14.2 |
| chr24_+_42127983 | 0.81 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr23_+_41200854 | 0.80 |
ENSDART00000109567
|
nhsa
|
Nance-Horan syndrome a (congenital cataracts and dental anomalies) |
| chr11_-_40504170 | 0.79 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr16_-_2870522 | 0.79 |
ENSDART00000148543
|
cdcp1a
|
CUB domain containing protein 1a |
| chr1_-_44638058 | 0.78 |
ENSDART00000081835
|
slc43a1b
|
solute carrier family 43 (amino acid system L transporter), member 1b |
| chr5_+_72152813 | 0.78 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr14_+_33264303 | 0.78 |
ENSDART00000130680
ENSDART00000075187 |
pdzd11
|
PDZ domain containing 11 |
| chr15_-_47857687 | 0.76 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr18_+_44649804 | 0.76 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr3_+_29469283 | 0.76 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr22_+_1940595 | 0.75 |
ENSDART00000163506
|
znf1167
|
zinc finger protein 1167 |
| chr16_-_1757521 | 0.75 |
ENSDART00000124660
|
ascc3
|
activating signal cointegrator 1 complex subunit 3 |
| chr6_-_13408680 | 0.74 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr18_-_41161828 | 0.74 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr20_-_7080427 | 0.74 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr2_-_6051836 | 0.73 |
ENSDART00000092479
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr4_-_23839789 | 0.72 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr5_+_61361815 | 0.72 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr15_-_20731297 | 0.72 |
ENSDART00000114464
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.6 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 2.9 | 8.7 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.0 | 4.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.0 | 4.9 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.7 | 3.5 | GO:1901166 | mesenchymal to epithelial transition(GO:0060231) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.6 | 1.9 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.6 | 3.0 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.6 | 10.3 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.6 | 2.4 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.6 | 1.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.6 | 2.8 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.5 | 1.5 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.5 | 1.9 | GO:0006272 | leading strand elongation(GO:0006272) nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.5 | 2.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.4 | 2.2 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.4 | 1.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 8.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 4.8 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.3 | 2.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.3 | 2.7 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.3 | 1.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 0.9 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.3 | 0.8 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.3 | 1.8 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.3 | 1.0 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.3 | 4.8 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.2 | 3.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.2 | 1.8 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 1.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 1.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.2 | 1.5 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 3.8 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.2 | 3.0 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 1.6 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.2 | 2.3 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 1.4 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 2.5 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 1.0 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.2 | 1.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 2.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.2 | 0.8 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.0 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.6 | GO:1903589 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.1 | 1.0 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.1 | 1.0 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 3.6 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 1.0 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 2.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.3 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 1.2 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.6 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.1 | 3.4 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.1 | 1.7 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.5 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 0.7 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.1 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.9 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.1 | 2.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.1 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.6 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 1.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.2 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.6 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.6 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 1.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 0.1 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 1.7 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 2.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.1 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.4 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.1 | 1.0 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.3 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.6 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.7 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 2.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 1.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 3.0 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 3.0 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 4.3 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 17.1 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 1.6 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.6 | GO:2000779 | regulation of double-strand break repair(GO:2000779) |
| 0.0 | 2.1 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.0 | 0.8 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 1.1 | GO:0050881 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 1.8 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 3.9 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.8 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 2.1 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.4 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 1.6 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 1.6 | GO:0006898 | receptor-mediated endocytosis(GO:0006898) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.8 | 3.0 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.6 | 3.9 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.5 | 1.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.4 | 1.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.3 | 1.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.3 | 1.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 0.9 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.3 | 1.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 2.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.8 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.7 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.8 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.8 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 1.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.7 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 1.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 3.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 17.2 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 3.6 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 4.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 10.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 2.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 5.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 3.2 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 1.0 | 3.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.0 | 3.9 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.9 | 10.3 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.9 | 2.7 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.6 | 8.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.6 | 1.7 | GO:0070905 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.5 | 4.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 1.9 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.4 | 8.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 2.8 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.4 | 2.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 1.5 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.3 | 1.6 | GO:2001070 | starch binding(GO:2001070) |
| 0.3 | 0.9 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.3 | 2.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 9.8 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.3 | 0.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.3 | 1.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.3 | 1.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.3 | 1.9 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.3 | 1.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.3 | 0.8 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 5.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 0.7 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.2 | 1.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 1.5 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 1.6 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.2 | 1.7 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 1.4 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.2 | 3.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 2.0 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.2 | 1.8 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.6 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 1.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 2.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 1.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 16.6 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.5 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 1.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 3.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 2.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 1.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 1.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 1.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.3 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.9 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.7 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.9 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 1.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 4.4 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.2 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 2.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.9 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.6 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 5.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 2.2 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 2.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 0.8 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.3 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.8 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 1.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 1.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 2.9 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 2.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 3.9 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.3 | 5.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.2 | 3.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.9 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.1 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 11.3 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 0.5 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.9 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 3.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 2.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 3.0 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.6 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 1.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.6 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |