Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for sp1
Z-value: 2.01
Transcription factors associated with sp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp1
|
ENSDARG00000088347 | sp1 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp1 | dr11_v1_chr11_-_28747_28747 | 0.75 | 2.1e-04 | Click! |
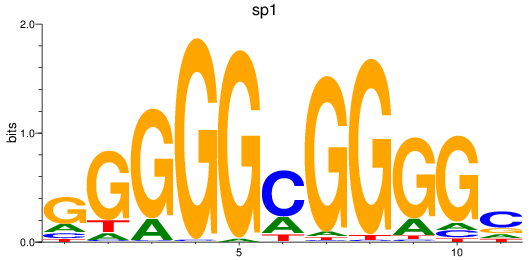
Activity profile of sp1 motif
Sorted Z-values of sp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_46201008 | 8.62 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr1_-_59571758 | 4.91 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr10_-_1625080 | 4.03 |
ENSDART00000137285
|
nup155
|
nucleoporin 155 |
| chr7_+_7630409 | 3.89 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr6_+_27991943 | 3.65 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr21_-_308852 | 3.37 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr24_+_17005647 | 3.07 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr6_+_39370587 | 2.99 |
ENSDART00000157165
ENSDART00000155079 |
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr24_-_37955993 | 2.93 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr8_-_20138054 | 2.92 |
ENSDART00000133141
ENSDART00000147634 ENSDART00000029939 |
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr3_+_1211242 | 2.87 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_+_46294337 | 2.85 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr12_-_43664682 | 2.83 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr9_+_15890558 | 2.80 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr1_-_51157454 | 2.76 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr25_+_16356083 | 2.68 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr16_-_7793457 | 2.67 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr16_-_7443388 | 2.65 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr1_-_59232267 | 2.65 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr5_-_66028371 | 2.61 |
ENSDART00000183012
|
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr24_-_41180149 | 2.52 |
ENSDART00000019975
|
acvr2ba
|
activin A receptor type 2Ba |
| chr3_+_41917499 | 2.51 |
ENSDART00000028673
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr18_-_49318823 | 2.50 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr2_-_48826707 | 2.46 |
ENSDART00000134711
|
svilb
|
supervillin b |
| chr9_-_46415847 | 2.43 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr5_+_43870389 | 2.42 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr20_+_33294428 | 2.41 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr7_-_16598212 | 2.41 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr15_+_1397811 | 2.37 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr1_-_51157660 | 2.27 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr20_+_39250673 | 2.26 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr3_-_53559408 | 2.22 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr22_-_34979139 | 2.20 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr23_-_3759345 | 2.18 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr25_-_36492779 | 2.17 |
ENSDART00000042271
|
irx3b
|
iroquois homeobox 3b |
| chr23_-_3759692 | 2.13 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr22_+_465269 | 2.11 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr22_-_26524596 | 2.09 |
ENSDART00000087623
|
zgc:194330
|
zgc:194330 |
| chr17_-_114121 | 2.07 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr13_-_33822550 | 2.06 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr1_+_57371447 | 2.04 |
ENSDART00000152229
ENSDART00000181077 |
si:dkey-27j5.3
|
si:dkey-27j5.3 |
| chr13_+_11439486 | 2.03 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr1_-_54107321 | 2.01 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr5_-_66028714 | 1.98 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr7_-_24995631 | 1.98 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr7_-_61683417 | 1.96 |
ENSDART00000098622
ENSDART00000184088 ENSDART00000148270 |
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr6_-_39313027 | 1.96 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr2_+_58841181 | 1.96 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr8_-_2591654 | 1.94 |
ENSDART00000049109
|
seta
|
SET nuclear proto-oncogene a |
| chr7_+_34688527 | 1.93 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr7_+_74141297 | 1.92 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr2_+_24374669 | 1.91 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr5_-_12219572 | 1.90 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr5_+_25765522 | 1.89 |
ENSDART00000133217
|
tmem2
|
transmembrane protein 2 |
| chr5_+_25760112 | 1.88 |
ENSDART00000088011
ENSDART00000182046 |
tmem2
|
transmembrane protein 2 |
| chr18_-_127558 | 1.88 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr3_-_53559581 | 1.88 |
ENSDART00000183499
|
notch3
|
notch 3 |
| chr3_+_15805917 | 1.85 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr2_+_26655744 | 1.85 |
ENSDART00000174928
|
asph
|
aspartate beta-hydroxylase |
| chr8_-_53490376 | 1.83 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr7_+_49715750 | 1.83 |
ENSDART00000019446
|
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr20_+_2731436 | 1.81 |
ENSDART00000058779
ENSDART00000129870 ENSDART00000132186 ENSDART00000152727 |
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr5_-_8171625 | 1.81 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr5_-_4532516 | 1.80 |
ENSDART00000192398
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr5_-_54714789 | 1.80 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr20_+_30797329 | 1.80 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr19_-_2029777 | 1.79 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr21_+_249970 | 1.79 |
ENSDART00000169026
|
jak2a
|
Janus kinase 2a |
| chr6_+_12922002 | 1.76 |
ENSDART00000080350
ENSDART00000149018 |
cxcr4a
|
chemokine (C-X-C motif) receptor 4a |
| chr10_-_31563049 | 1.72 |
ENSDART00000023575
|
robo3
|
roundabout, axon guidance receptor, homolog 3 (Drosophila) |
| chr18_-_46010 | 1.72 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr8_-_30979494 | 1.71 |
ENSDART00000138959
|
si:ch211-251j10.3
|
si:ch211-251j10.3 |
| chr18_-_127873 | 1.71 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr6_-_45905746 | 1.71 |
ENSDART00000025428
|
epha2a
|
eph receptor A2 a |
| chr22_+_13886821 | 1.70 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr8_+_2656681 | 1.70 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr1_+_29858032 | 1.70 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr11_+_31324335 | 1.69 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr2_-_39675829 | 1.68 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr23_-_2513300 | 1.68 |
ENSDART00000067652
|
snai1b
|
snail family zinc finger 1b |
| chr3_-_62527675 | 1.67 |
ENSDART00000155048
ENSDART00000064500 |
sox9b
|
SRY (sex determining region Y)-box 9b |
| chr24_-_21258945 | 1.67 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr3_+_12554801 | 1.66 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr8_-_51753604 | 1.66 |
ENSDART00000007090
|
tbx16
|
T-box 16 |
| chr25_+_3294150 | 1.65 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr12_-_37449396 | 1.64 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr20_-_16156419 | 1.63 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr9_+_34350025 | 1.63 |
ENSDART00000183451
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr6_-_27891961 | 1.61 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr7_-_52498175 | 1.60 |
ENSDART00000129769
|
cgnl1
|
cingulin-like 1 |
| chr25_-_37331513 | 1.59 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr3_-_5964557 | 1.59 |
ENSDART00000184738
|
BX284638.1
|
|
| chr20_+_48803248 | 1.59 |
ENSDART00000164006
|
nkx2.4b
|
NK2 homeobox 4b |
| chr5_-_44496805 | 1.59 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr5_-_14326959 | 1.59 |
ENSDART00000137355
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr16_-_52733384 | 1.57 |
ENSDART00000147236
ENSDART00000056101 |
azin1a
|
antizyme inhibitor 1a |
| chr1_-_23595779 | 1.56 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr21_-_11646878 | 1.56 |
ENSDART00000162426
ENSDART00000135937 ENSDART00000131448 ENSDART00000148097 ENSDART00000133443 |
cast
|
calpastatin |
| chr12_-_26851726 | 1.54 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr7_-_5396154 | 1.54 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr5_+_72152813 | 1.51 |
ENSDART00000149910
|
abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr3_-_21094437 | 1.50 |
ENSDART00000153739
ENSDART00000109790 |
nlk1
|
nemo-like kinase, type 1 |
| chr19_-_15855427 | 1.49 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr13_+_15941850 | 1.49 |
ENSDART00000016294
|
fignl1
|
fidgetin-like 1 |
| chr4_-_56703300 | 1.47 |
ENSDART00000160063
|
znf1118
|
zinc finger protein 1118 |
| chr2_+_24376373 | 1.47 |
ENSDART00000135323
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr17_-_10059557 | 1.47 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr10_+_10210455 | 1.47 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr20_-_28698172 | 1.46 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr3_+_46559639 | 1.46 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr7_+_1579236 | 1.46 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr13_-_40316367 | 1.45 |
ENSDART00000009343
|
pyroxd2
|
pyridine nucleotide-disulphide oxidoreductase domain 2 |
| chr25_-_348784 | 1.45 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr1_+_15062 | 1.44 |
ENSDART00000169005
|
cep97
|
centrosomal protein 97 |
| chr11_-_19775182 | 1.44 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr6_+_32326074 | 1.43 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr7_-_33114884 | 1.43 |
ENSDART00000052385
|
tph1b
|
tryptophan hydroxylase 1b |
| chr24_+_42127983 | 1.43 |
ENSDART00000190157
ENSDART00000176032 ENSDART00000175790 |
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr15_-_3736773 | 1.43 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr21_-_13972745 | 1.40 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr14_+_31865099 | 1.40 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr2_+_24374305 | 1.40 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr7_-_45852270 | 1.40 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr19_-_6193067 | 1.39 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr10_-_35257458 | 1.39 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr12_-_42214 | 1.39 |
ENSDART00000045071
|
foxk2
|
forkhead box K2 |
| chr20_+_710052 | 1.38 |
ENSDART00000166656
|
senp6a
|
SUMO1/sentrin specific peptidase 6a |
| chr5_-_54714525 | 1.37 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr4_-_5795309 | 1.37 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr10_-_641609 | 1.37 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr2_+_43469241 | 1.37 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr22_+_413349 | 1.37 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr14_+_31865324 | 1.37 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr23_+_21380079 | 1.37 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr23_+_5524247 | 1.36 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr7_-_18881358 | 1.36 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr14_-_12822 | 1.36 |
ENSDART00000180650
ENSDART00000188819 |
msx1a
|
muscle segment homeobox 1a |
| chr16_-_52646789 | 1.36 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr17_-_28811747 | 1.36 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr19_+_42716542 | 1.35 |
ENSDART00000144557
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr18_-_50845804 | 1.34 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr1_-_669717 | 1.34 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr14_-_42231293 | 1.34 |
ENSDART00000185486
|
BX890543.1
|
|
| chr22_+_10232527 | 1.33 |
ENSDART00000139297
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr15_+_19838458 | 1.33 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr5_+_68807170 | 1.33 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr1_+_14454663 | 1.31 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr4_-_23839789 | 1.31 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr19_+_48060464 | 1.31 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr5_-_63515210 | 1.30 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr21_-_37027252 | 1.30 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr8_+_31016180 | 1.30 |
ENSDART00000130870
ENSDART00000143604 |
odf2b
|
outer dense fiber of sperm tails 2b |
| chr7_-_51102479 | 1.30 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr24_+_17140938 | 1.29 |
ENSDART00000149134
|
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr17_-_10059996 | 1.29 |
ENSDART00000166984
|
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr13_+_5978809 | 1.29 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr3_-_32362872 | 1.28 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr14_-_237130 | 1.28 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr21_-_39024754 | 1.28 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr3_+_23692462 | 1.28 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr8_+_4697764 | 1.27 |
ENSDART00000064197
|
cldn5a
|
claudin 5a |
| chr18_-_14677936 | 1.26 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr20_+_51061695 | 1.26 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr23_+_31107685 | 1.26 |
ENSDART00000103448
|
tbx18
|
T-box 18 |
| chr20_-_2584101 | 1.26 |
ENSDART00000141595
ENSDART00000135760 |
med23
|
mediator complex subunit 23 |
| chr5_+_35398745 | 1.25 |
ENSDART00000098010
|
ptger4b
|
prostaglandin E receptor 4 (subtype EP4) b |
| chr8_+_29962635 | 1.25 |
ENSDART00000007640
|
ptch1
|
patched 1 |
| chr20_-_25748407 | 1.24 |
ENSDART00000063152
|
chst14
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 14 |
| chr16_-_53919115 | 1.24 |
ENSDART00000179533
|
fzd1
|
frizzled class receptor 1 |
| chr1_-_45584407 | 1.23 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr20_+_4157815 | 1.23 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr2_-_44282796 | 1.23 |
ENSDART00000163040
ENSDART00000166923 ENSDART00000056372 ENSDART00000109251 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr11_-_30138299 | 1.23 |
ENSDART00000172106
|
scml2
|
Scm polycomb group protein like 2 |
| chr5_+_67390645 | 1.23 |
ENSDART00000014822
|
ebf2
|
early B cell factor 2 |
| chr3_+_12322170 | 1.23 |
ENSDART00000161227
|
glis2b
|
GLIS family zinc finger 2b |
| chr12_+_48841182 | 1.22 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr7_-_64971839 | 1.22 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr12_+_14149686 | 1.22 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr23_+_44634187 | 1.21 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr7_+_22718251 | 1.21 |
ENSDART00000027718
ENSDART00000143341 |
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr25_+_150570 | 1.21 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr16_+_33953644 | 1.21 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr23_-_14403939 | 1.20 |
ENSDART00000090930
|
nkain4
|
sodium/potassium transporting ATPase interacting 4 |
| chr1_-_45616470 | 1.20 |
ENSDART00000150165
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_45580787 | 1.19 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr7_+_24881680 | 1.19 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr11_-_7139555 | 1.19 |
ENSDART00000016472
|
bmp7a
|
bone morphogenetic protein 7a |
| chr17_+_25856671 | 1.19 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr5_-_68782641 | 1.18 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr19_+_14352332 | 1.17 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr25_-_6389713 | 1.17 |
ENSDART00000083539
|
sin3aa
|
SIN3 transcription regulator family member Aa |
| chr23_+_10347851 | 1.17 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr10_+_17713022 | 1.17 |
ENSDART00000170453
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr4_-_1720648 | 1.16 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr6_+_54576520 | 1.16 |
ENSDART00000093199
ENSDART00000127519 ENSDART00000157142 |
tead3b
|
TEA domain family member 3 b |
| chr12_+_695619 | 1.16 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr17_+_51746830 | 1.16 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr16_+_32559821 | 1.15 |
ENSDART00000093250
|
pou3f2b
|
POU class 3 homeobox 2b |
| chr4_+_15006217 | 1.15 |
ENSDART00000090837
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr18_-_16924221 | 1.15 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 1.3 | 4.0 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 1.2 | 3.6 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.1 | 4.5 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.8 | 2.4 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.7 | 0.7 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.7 | 2.8 | GO:0048890 | epidermal cell fate specification(GO:0009957) lateral line ganglion development(GO:0048890) |
| 0.6 | 1.9 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.6 | 1.8 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.6 | 4.8 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.6 | 1.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.6 | 1.7 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) |
| 0.5 | 2.7 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.5 | 1.5 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.5 | 2.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.5 | 2.3 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.5 | 0.9 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.4 | 1.8 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.4 | 1.3 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.4 | 2.2 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.4 | 1.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.4 | 1.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.4 | 2.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.4 | 2.5 | GO:0055016 | hypochord development(GO:0055016) |
| 0.4 | 1.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.4 | 2.1 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 5.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.4 | 1.6 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.4 | 1.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.4 | 1.2 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.4 | 8.6 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.4 | 3.8 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.4 | 3.4 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.4 | 1.1 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.4 | 1.1 | GO:0009397 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.4 | 1.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.4 | 4.7 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.4 | 2.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 3.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.3 | 6.6 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.3 | 1.4 | GO:0090230 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.3 | 1.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 2.7 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.3 | 1.7 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.3 | 1.0 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 1.0 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.3 | 8.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.3 | 1.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.3 | 0.6 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.3 | 1.6 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.3 | 1.3 | GO:0042308 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 2.8 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.3 | 0.9 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.3 | 0.9 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.3 | 0.9 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.3 | 0.9 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.3 | 3.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.3 | 1.1 | GO:1900136 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.3 | 0.8 | GO:0043525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.3 | 1.7 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.3 | 1.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 1.6 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.3 | 1.3 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.3 | 1.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.3 | 1.3 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.2 | 1.0 | GO:0060471 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.2 | 1.0 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 1.0 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.2 | 0.7 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.2 | 1.7 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 2.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.2 | 1.8 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.2 | 1.4 | GO:1900044 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.2 | 0.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 0.7 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.2 | 1.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.1 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 0.4 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.2 | 0.6 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.2 | 1.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 0.6 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.2 | 2.1 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.6 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 0.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.2 | 0.4 | GO:0071867 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.6 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.2 | 1.4 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.2 | 0.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.2 | 0.8 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.2 | 0.6 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.2 | 2.5 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 3.1 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 0.4 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.2 | 0.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 0.5 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 1.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.2 | 0.5 | GO:0001113 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 0.9 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 0.5 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.2 | 1.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.3 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.2 | 1.0 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 3.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.2 | 1.2 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 2.5 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.2 | 1.5 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.2 | 1.0 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.2 | 0.6 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 1.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 0.8 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.2 | 0.6 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.2 | 1.7 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.2 | 0.3 | GO:0038179 | neurotrophin signaling pathway(GO:0038179) nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.6 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.2 | 0.5 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.8 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 2.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.3 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.6 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.7 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.1 | 1.8 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.1 | 0.7 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 1.7 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 1.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.1 | 1.1 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.4 | GO:0090244 | Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 | 1.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.5 | GO:0021854 | hypothalamus development(GO:0021854) |
| 0.1 | 1.7 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.1 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.4 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 1.2 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) |
| 0.1 | 0.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.1 | 0.8 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.1 | 0.5 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 1.3 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.3 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.1 | 0.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 2.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.9 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.6 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 2.1 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.1 | 0.7 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.7 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.9 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 1.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.6 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.7 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.1 | 2.2 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.1 | 3.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.4 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.6 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 0.4 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 1.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 0.5 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.2 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 1.3 | GO:0043268 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 2.9 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 8.1 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 2.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.6 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.9 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.3 | GO:1904950 | negative regulation of protein transport(GO:0051224) negative regulation of establishment of protein localization(GO:1904950) |
| 0.1 | 2.1 | GO:0007379 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.1 | 0.5 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.8 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.9 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.3 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) positive regulation of Golgi to plasma membrane protein transport(GO:0042998) regulation of establishment of protein localization to plasma membrane(GO:0090003) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.1 | 0.7 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.3 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 0.9 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 0.6 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 0.5 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.7 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.3 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.1 | 0.4 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.5 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 1.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.2 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 0.4 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.6 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.1 | 0.9 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.1 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 1.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.1 | 4.3 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 0.3 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.1 | 0.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 1.4 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.1 | 1.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.1 | 2.6 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.1 | 3.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 0.3 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 2.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.1 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.7 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.3 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.9 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.4 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 1.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.4 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.3 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.1 | 1.8 | GO:0046427 | positive regulation of JAK-STAT cascade(GO:0046427) positive regulation of STAT cascade(GO:1904894) |
| 0.1 | 0.8 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 2.6 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 0.6 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.6 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.9 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.2 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.1 | 1.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 0.4 | GO:0044319 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.6 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.5 | GO:0006582 | melanin metabolic process(GO:0006582) |
| 0.1 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.5 | GO:0072160 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 1.4 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.6 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 2.0 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.1 | 0.6 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.9 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 0.4 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 0.2 | GO:0032602 | chemokine production(GO:0032602) toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.1 | 0.5 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 2.0 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.1 | 0.5 | GO:0048634 | regulation of muscle organ development(GO:0048634) |
| 0.1 | 0.5 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.4 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 1.0 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 1.5 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 1.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.6 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0060137 | parturition(GO:0007567) multi-multicellular organism process(GO:0044706) maternal process involved in parturition(GO:0060137) |
| 0.0 | 1.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.6 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0055021 | regulation of cardiac muscle tissue growth(GO:0055021) regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.8 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.9 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.8 | GO:0014020 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) tube closure(GO:0060606) |
| 0.0 | 0.2 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 1.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0046850 | regulation of tissue remodeling(GO:0034103) regulation of bone remodeling(GO:0046850) |
| 0.0 | 4.9 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 2.1 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 0.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 4.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.2 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 5.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.6 | GO:0071897 | DNA biosynthetic process(GO:0071897) |
| 0.0 | 1.9 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.8 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 1.3 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 1.5 | GO:0016331 | morphogenesis of embryonic epithelium(GO:0016331) |
| 0.0 | 0.3 | GO:0051785 | positive regulation of mitotic nuclear division(GO:0045840) positive regulation of nuclear division(GO:0051785) |
| 0.0 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0060872 | semicircular canal development(GO:0060872) |
| 0.0 | 0.2 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.6 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.7 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 0.8 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 3.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 2.7 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.0 | 0.3 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 1.2 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 1.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.6 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.0 | 0.7 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.7 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 1.0 | GO:0001935 | endothelial cell proliferation(GO:0001935) |
| 0.0 | 1.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0007257 | activation of JUN kinase activity(GO:0007257) regulation of JUN kinase activity(GO:0043506) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 2.5 | GO:0045732 | positive regulation of protein catabolic process(GO:0045732) |
| 0.0 | 1.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.4 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 1.0 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.2 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.7 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 0.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.3 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 1.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 1.1 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.4 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.0 | GO:0006699 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.0 | 0.1 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.6 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.3 | GO:0031076 | embryonic camera-type eye development(GO:0031076) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 0.9 | GO:0044344 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.0 | 0.2 | GO:0033628 | regulation of cell adhesion mediated by integrin(GO:0033628) |
| 0.0 | 0.6 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.9 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.8 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.7 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.1 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 2.7 | GO:1904888 | cranial skeletal system development(GO:1904888) |
| 0.0 | 0.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 1.2 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 2.5 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 3.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 0.1 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0030534 | adult behavior(GO:0030534) |
| 0.0 | 0.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.5 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.2 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.1 | GO:0030517 | negative regulation of axon extension(GO:0030517) negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.2 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.0 | 4.0 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.4 | 1.1 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.3 | 1.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 1.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.3 | 2.4 | GO:0035060 | brahma complex(GO:0035060) |
| 0.3 | 2.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 1.7 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.3 | 2.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 3.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 2.5 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 0.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.8 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.2 | 2.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 3.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.2 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 0.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 1.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 0.8 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.5 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.1 | 0.8 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 1.6 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 1.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.5 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 2.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.6 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.6 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 0.3 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 1.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 2.0 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.4 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 9.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 6.4 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.1 | 0.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 4.6 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 3.0 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 0.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 0.2 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 0.3 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 1.0 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.5 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.1 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.1 | 0.8 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 3.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 1.0 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.4 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.7 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.0 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 1.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 1.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 1.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 2.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 5.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.2 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.0 | 8.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 2.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 3.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.7 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.1 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 1.3 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.3 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 87.7 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.0 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.9 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.7 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.4 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.6 | 1.9 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.6 | 2.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.5 | 2.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.5 | 2.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.4 | 1.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.4 | 1.3 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 1.3 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.4 | 8.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 10.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.4 | 1.1 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.4 | 1.1 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.4 | 4.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 1.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.3 | 3.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.3 | 2.8 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.3 | 0.9 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.3 | 3.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.6 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.3 | 1.9 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 2.7 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 1.0 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.3 | 5.6 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 3.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 2.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 1.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.2 | 1.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.2 | 3.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.8 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 0.6 | GO:0032143 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 3.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 4.6 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 4.0 | GO:0048185 | activin binding(GO:0048185) |
| 0.2 | 1.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 0.8 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 0.9 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 1.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 1.8 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 0.7 | GO:0008459 | chondroitin 6-sulfotransferase activity(GO:0008459) |
| 0.2 | 1.4 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 0.5 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.2 | 1.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.6 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 1.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 1.0 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.1 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 4.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 0.4 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.1 | 0.5 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.1 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.8 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.9 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 1.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 6.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 0.7 | GO:0042974 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.1 | 1.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 3.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 0.5 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.9 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 1.7 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.7 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 1.3 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.7 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.9 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 1.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.8 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 5.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.4 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 0.6 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.6 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.4 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.6 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.7 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 1.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 1.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.4 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.1 | 0.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.6 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 3.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 1.2 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 4.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 0.4 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 0.3 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.1 | 0.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.6 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.1 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 8.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.2 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 1.6 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.0 | 0.2 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 7.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.2 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.4 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.9 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.3 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 3.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.9 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 11.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 6.6 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.4 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.3 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 6.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.6 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 49.1 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.6 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 1.3 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.4 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 8.8 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.9 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.0 | 0.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 6.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.7 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.7 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 2.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 1.3 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.0 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 6.4 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 8.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 4.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 4.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 3.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 1.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 1.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 3.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 2.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.6 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 2.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 1.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 2.0 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.5 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 6.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 1.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 1.0 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.3 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID AVB3 INTEGRIN PATHWAY | Integrins in angiogenesis |
| 0.0 | 0.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.7 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.0 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.2 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.3 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 0.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.5 | 4.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.4 | 4.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.3 | 0.6 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 0.9 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.2 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 1.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.2 | 0.9 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 1.9 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 4.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.2 | 2.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 1.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 5.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 2.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.1 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.1 | 4.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 2.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.6 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.8 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 2.9 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.1 | 1.0 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 0.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.6 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 0.4 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.1 | 1.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 3.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 1.1 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 5.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.7 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 1.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 3.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 2.9 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 3.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.9 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.1 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.5 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME TRANSLATION | Genes involved in Translation |