Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
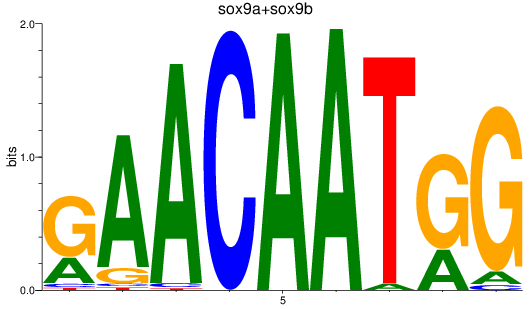
Results for sox9a+sox9b
Z-value: 1.24
Transcription factors associated with sox9a+sox9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox9a
|
ENSDARG00000003293 | SRY-box transcription factor 9a |
|
sox9b
|
ENSDARG00000043923 | SRY-box transcription factor 9b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox9b | dr11_v1_chr3_-_62527675_62527696 | 0.52 | 2.3e-02 | Click! |
| sox9a | dr11_v1_chr12_-_1951233_1951284 | -0.42 | 7.7e-02 | Click! |
Activity profile of sox9a+sox9b motif
Sorted Z-values of sox9a+sox9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_26704829 | 4.54 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr6_-_43092175 | 2.93 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr15_-_4528326 | 2.40 |
ENSDART00000158122
ENSDART00000155619 ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr23_-_21463788 | 2.31 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr23_+_21459263 | 2.09 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr9_-_46415847 | 1.95 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr14_-_41478265 | 1.86 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr23_+_35708730 | 1.74 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr22_-_10541372 | 1.74 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr16_+_35535375 | 1.67 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr4_+_5333988 | 1.58 |
ENSDART00000129398
ENSDART00000163850 ENSDART00000067374 ENSDART00000150780 ENSDART00000150493 ENSDART00000150306 |
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr23_-_43393083 | 1.53 |
ENSDART00000112758
|
zgc:174862
|
zgc:174862 |
| chr8_-_46897734 | 1.47 |
ENSDART00000138125
|
hes2.2
|
hes family bHLH transcription factor 2, tandem duplicate 2 |
| chr1_+_218524 | 1.46 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr4_+_5334439 | 1.46 |
ENSDART00000180644
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr16_+_35535171 | 1.46 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr6_+_12968101 | 1.45 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr19_-_41472228 | 1.43 |
ENSDART00000113388
|
dlx5a
|
distal-less homeobox 5a |
| chr22_+_15960005 | 1.41 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr7_+_19850889 | 1.41 |
ENSDART00000100782
|
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr22_+_15960514 | 1.37 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr12_-_28848015 | 1.35 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr15_+_5973909 | 1.34 |
ENSDART00000126886
ENSDART00000189618 |
igsf5b
|
immunoglobulin superfamily, member 5b |
| chr14_-_33978117 | 1.34 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr3_+_52999962 | 1.33 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr22_+_15959844 | 1.33 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr19_+_19777437 | 1.32 |
ENSDART00000170662
|
hoxa3a
|
homeobox A3a |
| chr24_-_41797681 | 1.31 |
ENSDART00000169643
|
arhgap28
|
Rho GTPase activating protein 28 |
| chr10_-_28027490 | 1.31 |
ENSDART00000185445
|
ints2
|
integrator complex subunit 2 |
| chr13_-_35908275 | 1.27 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr14_+_16287968 | 1.26 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr3_-_9444749 | 1.25 |
ENSDART00000182191
|
FO904885.3
|
|
| chr8_+_15254564 | 1.24 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr16_-_42461263 | 1.23 |
ENSDART00000109259
|
smarcc1a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
| chr2_+_11685742 | 1.21 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr10_-_28761454 | 1.16 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr23_-_21453614 | 1.14 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr2_+_58841181 | 1.11 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr15_-_5624361 | 1.09 |
ENSDART00000176446
ENSDART00000114410 |
wdr62
|
WD repeat domain 62 |
| chr7_-_17028015 | 1.09 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr13_-_35907768 | 1.09 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr7_-_18601206 | 1.09 |
ENSDART00000111636
|
DTX4
|
si:ch211-119e14.2 |
| chr5_+_25762271 | 1.08 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr15_-_35246742 | 1.08 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr4_+_5334202 | 1.07 |
ENSDART00000150409
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr14_+_38786298 | 1.07 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr1_-_8651718 | 1.06 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr3_-_26805455 | 1.05 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr16_+_54209504 | 1.05 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr9_+_2002701 | 1.04 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr5_-_22082918 | 1.03 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr9_+_32978302 | 1.03 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr17_-_42097267 | 1.02 |
ENSDART00000110871
ENSDART00000155484 |
nkx2.4a
|
NK2 homeobox 4a |
| chr19_+_47394270 | 1.02 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr23_-_30727596 | 1.00 |
ENSDART00000060193
|
thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr5_-_22130937 | 1.00 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr9_-_2594410 | 1.00 |
ENSDART00000188306
ENSDART00000164276 |
sp9
|
sp9 transcription factor |
| chr5_-_14344647 | 1.00 |
ENSDART00000188456
|
tet3
|
tet methylcytosine dioxygenase 3 |
| chr18_+_20034023 | 1.00 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr22_-_10570749 | 1.00 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr23_+_8797143 | 1.00 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr19_-_18418763 | 0.99 |
ENSDART00000167271
|
zgc:112966
|
zgc:112966 |
| chr18_+_48428713 | 0.99 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr13_+_51579851 | 0.99 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr3_+_17653784 | 0.97 |
ENSDART00000159984
ENSDART00000157682 ENSDART00000187937 |
kat2a
|
K(lysine) acetyltransferase 2A |
| chr7_+_22801465 | 0.97 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr16_+_33121260 | 0.97 |
ENSDART00000058472
|
akirin1
|
akirin 1 |
| chr16_+_33121106 | 0.97 |
ENSDART00000110195
|
akirin1
|
akirin 1 |
| chr9_+_34950942 | 0.95 |
ENSDART00000077800
|
tfdp1a
|
transcription factor Dp-1, a |
| chr21_+_19547806 | 0.95 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr23_-_33944597 | 0.94 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr23_+_38251864 | 0.94 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr20_+_26943072 | 0.93 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr7_-_24621689 | 0.93 |
ENSDART00000101265
|
pik3c3
|
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr4_-_56898328 | 0.92 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr4_-_211714 | 0.92 |
ENSDART00000172566
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr3_+_23743139 | 0.92 |
ENSDART00000187409
|
hoxb3a
|
homeobox B3a |
| chr23_-_27235403 | 0.91 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr15_+_14592624 | 0.91 |
ENSDART00000162350
|
FBXO46
|
si:dkey-114g7.4 |
| chr17_+_132555 | 0.91 |
ENSDART00000158159
|
zgc:77287
|
zgc:77287 |
| chr9_+_35016201 | 0.91 |
ENSDART00000182404
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr6_-_33875919 | 0.91 |
ENSDART00000190411
|
tmem69
|
transmembrane protein 69 |
| chr18_-_45761868 | 0.91 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr5_-_32363372 | 0.90 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr20_+_54290356 | 0.89 |
ENSDART00000173347
|
zp2.2
|
zona pellucida glycoprotein 2, tandem duplicate 2 |
| chr5_+_36900157 | 0.89 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr10_-_28028998 | 0.89 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr11_+_25328199 | 0.88 |
ENSDART00000141478
ENSDART00000112209 |
fam83d
|
family with sequence similarity 83, member D |
| chr21_-_37194365 | 0.87 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr16_+_52847079 | 0.86 |
ENSDART00000163151
|
cep72
|
centrosomal protein 72 |
| chr4_-_1684335 | 0.86 |
ENSDART00000019144
ENSDART00000113360 |
arid2
|
AT rich interactive domain 2 (ARID, RFX-like) |
| chr16_-_12060488 | 0.86 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr11_+_39135050 | 0.85 |
ENSDART00000180571
ENSDART00000189685 |
cdc42
|
cell division cycle 42 |
| chr10_+_9561066 | 0.85 |
ENSDART00000136281
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr3_+_23742868 | 0.85 |
ENSDART00000153512
|
hoxb3a
|
homeobox B3a |
| chr15_-_47865063 | 0.84 |
ENSDART00000151600
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr21_-_37194669 | 0.84 |
ENSDART00000192748
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr16_+_25074029 | 0.84 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr25_+_18475032 | 0.83 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr5_-_67365006 | 0.83 |
ENSDART00000136116
|
unga
|
uracil DNA glycosylase a |
| chr7_-_24995631 | 0.83 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr20_-_3319642 | 0.83 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr12_+_33460794 | 0.83 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr17_-_36936649 | 0.81 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr13_+_47050726 | 0.81 |
ENSDART00000140045
|
anapc1
|
anaphase promoting complex subunit 1 |
| chr20_+_13141408 | 0.80 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr2_+_21634128 | 0.80 |
ENSDART00000089822
|
fbxl7
|
F-box and leucine-rich repeat protein 7 |
| chr19_-_7540821 | 0.80 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr21_-_25565392 | 0.80 |
ENSDART00000144917
ENSDART00000180102 |
si:dkey-17e16.10
|
si:dkey-17e16.10 |
| chr17_-_20228610 | 0.80 |
ENSDART00000125758
|
ebf3b
|
early B cell factor 3b |
| chr19_+_19762183 | 0.80 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr21_-_37194839 | 0.80 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr17_+_23968214 | 0.79 |
ENSDART00000183053
|
xpo1b
|
exportin 1 (CRM1 homolog, yeast) b |
| chr4_-_2059233 | 0.79 |
ENSDART00000188177
ENSDART00000129521 ENSDART00000082289 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr6_-_34220641 | 0.79 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr3_-_5644028 | 0.79 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr3_+_53156813 | 0.78 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr24_-_2381143 | 0.78 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr12_+_27536095 | 0.78 |
ENSDART00000013033
|
etv4
|
ets variant 4 |
| chr3_-_16110351 | 0.77 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr22_-_21046654 | 0.77 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr12_-_10476448 | 0.77 |
ENSDART00000106172
|
rac1a
|
Rac family small GTPase 1a |
| chr25_+_35502552 | 0.76 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr14_+_24277556 | 0.76 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr14_-_36397768 | 0.74 |
ENSDART00000185199
ENSDART00000052562 |
spata4
|
spermatogenesis associated 4 |
| chr7_+_38811800 | 0.74 |
ENSDART00000052322
|
zgc:110699
|
zgc:110699 |
| chr7_-_22941472 | 0.74 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr6_+_16031189 | 0.73 |
ENSDART00000015333
|
gbx2
|
gastrulation brain homeobox 2 |
| chr25_-_14433503 | 0.73 |
ENSDART00000103957
|
exoc3l1
|
exocyst complex component 3-like 1 |
| chr12_+_27536270 | 0.73 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr20_+_1272526 | 0.73 |
ENSDART00000008115
ENSDART00000133825 |
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr15_+_23722620 | 0.73 |
ENSDART00000011447
|
sae1
|
SUMO1 activating enzyme subunit 1 |
| chr25_+_3294150 | 0.73 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr8_-_19342111 | 0.73 |
ENSDART00000138881
|
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr23_-_33738945 | 0.73 |
ENSDART00000136386
|
si:ch211-210c8.7
|
si:ch211-210c8.7 |
| chr4_-_44872722 | 0.73 |
ENSDART00000150621
|
BX649307.1
|
|
| chr20_+_54295213 | 0.72 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr13_-_11035420 | 0.71 |
ENSDART00000108709
|
cep170aa
|
centrosomal protein 170Aa |
| chr16_+_12517535 | 0.71 |
ENSDART00000155407
|
rasip1
|
Ras interacting protein 1 |
| chr7_-_33023404 | 0.71 |
ENSDART00000052383
|
cd81a
|
CD81 molecule a |
| chr2_+_26303627 | 0.71 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr21_-_11054876 | 0.71 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr20_+_54304800 | 0.71 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr7_+_25221757 | 0.71 |
ENSDART00000173551
|
exoc6b
|
exocyst complex component 6B |
| chr10_-_25628555 | 0.70 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr10_-_17587832 | 0.70 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr5_+_24086227 | 0.70 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr3_+_32410746 | 0.70 |
ENSDART00000025496
|
rras
|
RAS related |
| chr23_+_44236281 | 0.70 |
ENSDART00000149842
|
MEPCE
|
si:ch1073-157b13.1 |
| chr8_+_46641314 | 0.69 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr7_+_21180747 | 0.69 |
ENSDART00000185543
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr7_-_30624435 | 0.69 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr25_-_13789955 | 0.69 |
ENSDART00000167742
ENSDART00000165116 ENSDART00000171461 |
ckap5
|
cytoskeleton associated protein 5 |
| chr17_+_15535501 | 0.68 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr22_-_12763350 | 0.68 |
ENSDART00000143574
|
cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr25_+_27405738 | 0.68 |
ENSDART00000183266
ENSDART00000115139 |
pot1
|
protection of telomeres 1 homolog |
| chr22_+_1911269 | 0.67 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr19_+_37620342 | 0.67 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr18_-_16924221 | 0.67 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr9_+_37152564 | 0.67 |
ENSDART00000189497
|
gli2a
|
GLI family zinc finger 2a |
| chr16_-_9694822 | 0.67 |
ENSDART00000168748
|
COLEC10
|
si:ch211-93i7.4 |
| chr2_-_5135125 | 0.67 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr3_-_32873641 | 0.67 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr22_-_21046843 | 0.66 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr20_+_54309148 | 0.66 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr17_-_24521382 | 0.66 |
ENSDART00000092948
|
peli1b
|
pellino E3 ubiquitin protein ligase 1b |
| chr17_+_38602790 | 0.65 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr21_+_19330774 | 0.65 |
ENSDART00000109412
|
helq
|
helicase, POLQ like |
| chr2_-_21167652 | 0.64 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr17_-_36936856 | 0.64 |
ENSDART00000010274
ENSDART00000188887 |
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr5_-_36949476 | 0.64 |
ENSDART00000047269
|
h3f3c
|
H3 histone, family 3C |
| chr1_+_52792439 | 0.64 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr24_+_17142881 | 0.64 |
ENSDART00000177272
ENSDART00000192259 ENSDART00000191029 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr3_+_31058464 | 0.64 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr2_-_22286828 | 0.64 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr16_+_14710436 | 0.63 |
ENSDART00000027982
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr2_-_26596794 | 0.63 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr3_-_45778123 | 0.63 |
ENSDART00000146211
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr7_+_24006875 | 0.63 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr9_-_22963897 | 0.63 |
ENSDART00000133676
|
si:dkey-91i10.2
|
si:dkey-91i10.2 |
| chr19_+_7549854 | 0.63 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr13_+_35746440 | 0.63 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr19_+_14059349 | 0.63 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr7_+_24390939 | 0.63 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr23_+_44349252 | 0.62 |
ENSDART00000097644
|
ephb4b
|
eph receptor B4b |
| chr25_+_18965430 | 0.62 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr3_+_29640996 | 0.62 |
ENSDART00000011052
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr8_+_48491387 | 0.62 |
ENSDART00000086053
|
PRDM16
|
si:ch211-263k4.2 |
| chr24_+_13635108 | 0.62 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr8_-_32385989 | 0.62 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr23_+_35650771 | 0.62 |
ENSDART00000005158
|
ccnt1
|
cyclin T1 |
| chr4_-_56068511 | 0.62 |
ENSDART00000168345
|
znf1133
|
zinc finger protein 1133 |
| chr3_+_32689707 | 0.61 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr17_+_24597001 | 0.61 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr3_+_34234029 | 0.61 |
ENSDART00000044859
|
znf207a
|
zinc finger protein 207, a |
| chr5_+_60590796 | 0.61 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr3_-_37148594 | 0.60 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr21_-_41870029 | 0.60 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr12_-_37449396 | 0.60 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr2_+_32846602 | 0.60 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox9a+sox9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.8 | 2.5 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.5 | 4.4 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.5 | 1.9 | GO:0014857 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) positive regulation of lamellipodium organization(GO:1902745) |
| 0.5 | 4.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.3 | 2.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.3 | 2.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.3 | 0.8 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.3 | 0.8 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.3 | 1.0 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.3 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.2 | 5.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.2 | 0.7 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 4.8 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.2 | 1.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.3 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.2 | 0.6 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 1.0 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 0.6 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.5 | GO:0034473 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 2.9 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.2 | 1.8 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.2 | 1.0 | GO:0021767 | mammillary body development(GO:0021767) neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 0.8 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 0.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.2 | 0.9 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 1.5 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.1 | 0.6 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.1 | 0.4 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.1 | 0.7 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.7 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.7 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.0 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 2.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.1 | 0.5 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 1.0 | GO:0060173 | limb development(GO:0060173) |
| 0.1 | 0.4 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.4 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.5 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.1 | 0.3 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.9 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.8 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 1.1 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.8 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.1 | 1.6 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.1 | 0.1 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.1 | 1.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.1 | 2.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.7 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.1 | 0.4 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 0.7 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.6 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.3 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.5 | GO:0042664 | negative regulation of endodermal cell fate specification(GO:0042664) |
| 0.1 | 0.4 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.2 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.4 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.1 | 0.7 | GO:0021871 | forebrain regionalization(GO:0021871) |
| 0.1 | 0.7 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.5 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.1 | 0.9 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 0.7 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 | 0.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.1 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.1 | 0.3 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.5 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.2 | GO:2001014 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.2 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.7 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 2.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.1 | 0.6 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.6 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 1.1 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.1 | 0.7 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.1 | 0.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 0.3 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.2 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.4 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0042694 | muscle cell fate specification(GO:0042694) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.5 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.9 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.5 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 2.3 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.5 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.4 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 1.2 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.3 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.2 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 1.2 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.6 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.5 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 1.1 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 1.0 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 1.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.1 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 1.0 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.4 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.4 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.4 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.8 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.5 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 2.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.1 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.4 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:1902269 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.7 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.3 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.5 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 4.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.1 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.9 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.5 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.6 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0090307 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.0 | 0.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.4 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.6 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 1.1 | GO:0032880 | regulation of protein localization(GO:0032880) |
| 0.0 | 0.5 | GO:0050982 | detection of mechanical stimulus(GO:0050982) |
| 0.0 | 1.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.4 | GO:0031103 | axon regeneration(GO:0031103) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 0.8 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 0.8 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 1.4 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.2 | 1.3 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.6 | GO:0097361 | CIA complex(GO:0097361) |
| 0.2 | 0.9 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.2 | 0.7 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.2 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.4 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.5 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 2.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.1 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 1.7 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 2.0 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.1 | 0.4 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.1 | 1.0 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 0.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 1.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 5.9 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.5 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.6 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 0.4 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.1 | 0.4 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 1.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.5 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 5.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.8 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.5 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.3 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.0 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.7 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.8 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.6 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.3 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 1.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 5.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.2 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0044450 | microtubule organizing center part(GO:0044450) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.1 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.4 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.1 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.6 | 4.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 1.5 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.3 | 1.5 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 0.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.7 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.2 | 0.7 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.2 | 0.6 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.6 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.2 | 0.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 0.6 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 2.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.6 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 1.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.4 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.1 | 0.4 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 1.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 1.3 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.5 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 0.5 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.1 | 0.4 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.9 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 0.6 | GO:0008263 | mismatch base pair DNA N-glycosylase activity(GO:0000700) pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.8 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.5 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.6 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 0.7 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.3 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.5 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.4 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 1.5 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 1.1 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.4 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.6 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.0 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 0.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.6 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.7 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.2 | GO:0004984 | olfactory receptor activity(GO:0004984) odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 1.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 1.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 4.3 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.0 | 0.5 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.0 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 4.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.0 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 4.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 11.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 6.4 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 3.2 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.9 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 31.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.6 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.8 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.0 | 0.8 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.1 | 3.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 2.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 0.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 4.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 4.5 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 2.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 2.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 1.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.9 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.7 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.3 | 1.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.3 | 2.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 0.9 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 0.7 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.5 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 0.4 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.7 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.5 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.9 | REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | Genes involved in Recruitment of mitotic centrosome proteins and complexes |
| 0.0 | 1.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 2.4 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.0 | REACTOME FORMATION OF RNA POL II ELONGATION COMPLEX | Genes involved in Formation of RNA Pol II elongation complex |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.9 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.2 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |