Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
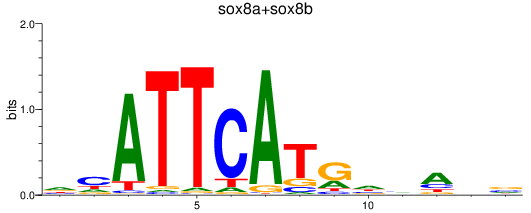
Results for sox8a+sox8b
Z-value: 0.62
Transcription factors associated with sox8a+sox8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox8b
|
ENSDARG00000037782 | SRY-box transcription factor 8b |
|
sox8a
|
ENSDARG00000105301 | SRY-box transcription factor 8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox8a | dr11_v1_chr12_-_1085227_1085227 | -0.69 | 1.1e-03 | Click! |
| sox8b | dr11_v1_chr3_-_62403550_62403550 | -0.20 | 4.0e-01 | Click! |
Activity profile of sox8a+sox8b motif
Sorted Z-values of sox8a+sox8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11949996 | 1.94 |
ENSDART00000163478
ENSDART00000167299 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr19_-_11950434 | 1.90 |
ENSDART00000024861
|
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr23_-_27235403 | 1.82 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr19_+_27448625 | 1.44 |
ENSDART00000052352
|
ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr9_+_34952203 | 1.44 |
ENSDART00000121828
ENSDART00000142347 |
tfdp1a
|
transcription factor Dp-1, a |
| chr3_-_27647845 | 1.35 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr11_-_30138299 | 1.33 |
ENSDART00000172106
|
scml2
|
Scm polycomb group protein like 2 |
| chr17_+_24687338 | 1.26 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr19_-_17208728 | 1.17 |
ENSDART00000151228
|
stmn1a
|
stathmin 1a |
| chr14_+_38786298 | 1.13 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr9_-_8979468 | 1.10 |
ENSDART00000134646
|
ing1
|
inhibitor of growth family, member 1 |
| chr21_+_34088110 | 1.08 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr12_+_30705769 | 1.07 |
ENSDART00000186448
ENSDART00000066259 |
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr24_-_21923930 | 1.05 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr22_+_9522971 | 1.05 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr11_-_18015534 | 1.02 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr4_+_22297839 | 0.99 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr2_+_24770435 | 0.95 |
ENSDART00000078854
|
mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr5_+_45008418 | 0.92 |
ENSDART00000189882
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr8_+_41238064 | 0.89 |
ENSDART00000111321
|
zgc:152830
|
zgc:152830 |
| chr2_-_24554416 | 0.89 |
ENSDART00000052061
|
cnn2
|
calponin 2 |
| chr13_-_44423 | 0.86 |
ENSDART00000093247
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr2_+_15048410 | 0.84 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr7_+_25003851 | 0.83 |
ENSDART00000144526
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr1_+_38362412 | 0.82 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr9_-_23147026 | 0.78 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr9_-_8979154 | 0.77 |
ENSDART00000145266
|
ing1
|
inhibitor of growth family, member 1 |
| chr21_+_42717424 | 0.74 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr1_-_26063188 | 0.72 |
ENSDART00000168640
|
pdcd4a
|
programmed cell death 4a |
| chr10_-_14545658 | 0.72 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr3_+_62327332 | 0.71 |
ENSDART00000158414
|
BX470259.1
|
|
| chr20_-_39391833 | 0.71 |
ENSDART00000135149
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr22_-_14262115 | 0.70 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr12_+_30706158 | 0.69 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr21_+_43328685 | 0.66 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr2_-_22492289 | 0.65 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr7_+_52753067 | 0.65 |
ENSDART00000053814
|
mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr21_+_34088377 | 0.64 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr14_+_48862987 | 0.63 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr21_+_39673157 | 0.61 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr19_-_27448606 | 0.61 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| chr6_-_48347724 | 0.60 |
ENSDART00000046973
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr23_-_36418708 | 0.59 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
| chr17_+_24604373 | 0.57 |
ENSDART00000082193
|
zmpste24
|
zinc metallopeptidase, STE24 homolog |
| chr10_+_11398111 | 0.56 |
ENSDART00000064210
|
cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr12_+_34258139 | 0.55 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr19_+_11432330 | 0.54 |
ENSDART00000188065
|
PQLC1
|
PQ loop repeat containing 1 |
| chr20_-_39391492 | 0.54 |
ENSDART00000184886
|
si:dkey-217m5.8
|
si:dkey-217m5.8 |
| chr10_+_7709724 | 0.52 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr13_+_8892784 | 0.51 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr1_-_16688965 | 0.50 |
ENSDART00000079001
ENSDART00000178599 |
frg1
|
FSHD region gene 1 |
| chr17_-_33707589 | 0.49 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr7_-_18547420 | 0.49 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr17_+_14784275 | 0.49 |
ENSDART00000005355
|
rtraf
|
RNA transcription, translation and transport factor |
| chr15_-_43327911 | 0.48 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr14_+_11457500 | 0.45 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr3_+_39579393 | 0.45 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr6_+_13506841 | 0.44 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr19_+_3653976 | 0.43 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr7_+_65121743 | 0.37 |
ENSDART00000108471
|
ipo7
|
importin 7 |
| chr25_-_37248795 | 0.37 |
ENSDART00000087247
ENSDART00000154045 |
glg1a
|
golgi glycoprotein 1a |
| chr21_-_43328056 | 0.36 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr10_+_5268054 | 0.36 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr16_-_25607266 | 0.35 |
ENSDART00000192602
|
zgc:110410
|
zgc:110410 |
| chr5_+_41146786 | 0.34 |
ENSDART00000175766
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr20_+_30578967 | 0.33 |
ENSDART00000010494
|
fgfr1op
|
FGFR1 oncogene partner |
| chr23_+_4890693 | 0.32 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr16_-_25606889 | 0.32 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr16_-_21047872 | 0.31 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr8_-_7603516 | 0.31 |
ENSDART00000179826
ENSDART00000190153 |
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr16_+_46410520 | 0.31 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr6_-_37743724 | 0.31 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr3_+_32663865 | 0.31 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr7_-_50410524 | 0.30 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr22_-_817479 | 0.30 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr8_-_7603700 | 0.30 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr8_-_46386024 | 0.29 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr10_+_40629616 | 0.29 |
ENSDART00000147476
|
CR396590.9
|
|
| chr15_+_28368823 | 0.29 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr12_+_18544159 | 0.28 |
ENSDART00000162900
|
mlst8
|
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr8_+_42718135 | 0.27 |
ENSDART00000158900
|
zgc:194007
|
zgc:194007 |
| chr14_-_25452503 | 0.26 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr11_+_13176568 | 0.26 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr13_+_30225262 | 0.24 |
ENSDART00000141945
|
rps24
|
ribosomal protein S24 |
| chr9_-_3934963 | 0.23 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr20_-_3276339 | 0.23 |
ENSDART00000166831
|
rps6kc1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr11_-_45138857 | 0.22 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr1_+_6646529 | 0.22 |
ENSDART00000144641
ENSDART00000103701 ENSDART00000138919 |
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr14_+_6546516 | 0.21 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr19_-_24163845 | 0.21 |
ENSDART00000133277
|
prf1.2
|
perforin 1.2 |
| chr6_+_49926115 | 0.20 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr3_-_3413669 | 0.17 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr19_-_24198139 | 0.16 |
ENSDART00000138864
ENSDART00000125350 |
prf1.7
|
perforin 1.7 |
| chr19_-_19664743 | 0.13 |
ENSDART00000187419
|
BX601646.1
|
|
| chr18_+_45666489 | 0.13 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr10_+_9595575 | 0.08 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr5_+_31214341 | 0.07 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr16_+_25095483 | 0.07 |
ENSDART00000155032
|
si:ch211-261d7.3
|
si:ch211-261d7.3 |
| chr18_+_5917625 | 0.06 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr9_+_23714406 | 0.04 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr4_+_39766931 | 0.03 |
ENSDART00000138663
|
si:dkeyp-85d8.1
|
si:dkeyp-85d8.1 |
| chr23_-_35694171 | 0.02 |
ENSDART00000077539
|
tuba1c
|
tubulin, alpha 1c |
| chr9_+_50001746 | 0.02 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr3_+_3573696 | 0.01 |
ENSDART00000169680
|
CR589947.2
|
|
| chr16_+_40508882 | 0.01 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr4_+_6833583 | 0.01 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr22_+_24673168 | 0.00 |
ENSDART00000135257
|
si:rp71-23d18.8
|
si:rp71-23d18.8 |
| chr15_+_28368644 | 0.00 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox8a+sox8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 0.6 | GO:0080120 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.2 | 1.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 1.3 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.1 | 3.8 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 0.7 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.5 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 1.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.6 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.5 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 1.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.8 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.7 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.2 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.9 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 1.7 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.9 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.9 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.5 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.6 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.3 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.2 | 0.9 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.1 | 0.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 0.6 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.1 | 0.7 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.8 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.7 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.2 | 1.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 1.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.5 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.8 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 1.8 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.4 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.7 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.6 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.5 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 2.0 | GO:0005516 | calmodulin binding(GO:0005516) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.8 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.9 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |