Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
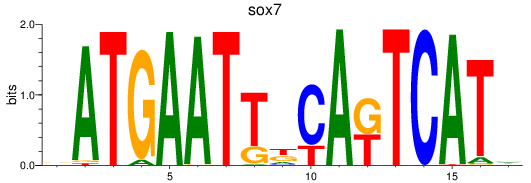
Results for sox7
Z-value: 0.57
Transcription factors associated with sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox7
|
ENSDARG00000030125 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox7 | dr11_v1_chr20_+_19066858_19066858 | 0.23 | 3.5e-01 | Click! |
Activity profile of sox7 motif
Sorted Z-values of sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_26535875 | 0.80 |
ENSDART00000135988
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr2_-_24996441 | 0.78 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr23_-_26536055 | 0.64 |
ENSDART00000182719
|
si:dkey-205h13.2
|
si:dkey-205h13.2 |
| chr9_-_23033818 | 0.56 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr11_+_10541258 | 0.55 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr5_+_34982318 | 0.53 |
ENSDART00000160504
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr8_+_23174137 | 0.53 |
ENSDART00000189470
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr25_-_22639133 | 0.52 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr20_+_50956369 | 0.49 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr15_-_21692630 | 0.49 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr21_-_18932761 | 0.48 |
ENSDART00000140129
|
med15
|
mediator complex subunit 15 |
| chr12_-_4540564 | 0.47 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr8_+_16726386 | 0.45 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr8_-_22514918 | 0.45 |
ENSDART00000021514
ENSDART00000189272 |
apex2
|
APEX nuclease (apurinic/apyrimidinic endonuclease) 2 |
| chr25_-_7999756 | 0.44 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr2_+_6991208 | 0.44 |
ENSDART00000182239
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr21_+_4320769 | 0.40 |
ENSDART00000168553
|
HTRA2 (1 of many)
|
si:dkey-84o3.4 |
| chr16_+_27345383 | 0.39 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr15_-_23647078 | 0.38 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr22_-_29191152 | 0.38 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr7_-_61288882 | 0.38 |
ENSDART00000098616
ENSDART00000137762 |
rwdd
|
RWD domain containing 4 |
| chr11_+_3308656 | 0.36 |
ENSDART00000082458
|
sarnp
|
SAP domain containing ribonucleoprotein |
| chr19_-_42462491 | 0.35 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr7_-_13648635 | 0.35 |
ENSDART00000148957
ENSDART00000130967 |
spg21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr16_-_31933740 | 0.33 |
ENSDART00000125411
|
si:ch1073-90m23.1
|
si:ch1073-90m23.1 |
| chr16_+_40024883 | 0.33 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr22_-_26558166 | 0.30 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr10_-_23379146 | 0.30 |
ENSDART00000056369
|
cadm2a
|
cell adhesion molecule 2a |
| chr20_+_38276690 | 0.29 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr17_+_15534815 | 0.29 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr21_-_21570134 | 0.27 |
ENSDART00000174388
|
or133-4
|
odorant receptor, family H, subfamily 133, member 4 |
| chr21_+_19062124 | 0.26 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr13_-_30161684 | 0.26 |
ENSDART00000040409
|
ppa1b
|
pyrophosphatase (inorganic) 1b |
| chr5_-_57169604 | 0.26 |
ENSDART00000136232
|
si:dkey-79c1.1
|
si:dkey-79c1.1 |
| chr16_+_46111849 | 0.25 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr15_-_35252522 | 0.24 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr1_-_10669436 | 0.23 |
ENSDART00000152421
|
si:dkey-31e10.5
|
si:dkey-31e10.5 |
| chr21_-_41147818 | 0.23 |
ENSDART00000167339
ENSDART00000192730 |
msx2b
|
muscle segment homeobox 2b |
| chr8_+_35664152 | 0.22 |
ENSDART00000144520
|
si:dkeyp-14d3.1
|
si:dkeyp-14d3.1 |
| chr21_-_19330333 | 0.22 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr13_+_835390 | 0.20 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr14_+_45645024 | 0.19 |
ENSDART00000168278
|
si:ch211-276i12.4
|
si:ch211-276i12.4 |
| chr25_+_28253844 | 0.19 |
ENSDART00000151891
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr6_-_16735402 | 0.19 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr22_-_26852516 | 0.18 |
ENSDART00000005829
|
gde1
|
glycerophosphodiester phosphodiesterase 1 |
| chr19_-_11031145 | 0.18 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr9_-_9989660 | 0.18 |
ENSDART00000081463
|
ugt1ab
|
UDP glucuronosyltransferase 1 family a, b |
| chr6_+_49081141 | 0.17 |
ENSDART00000131080
ENSDART00000050000 |
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr1_+_36674584 | 0.16 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr10_+_22034477 | 0.16 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr8_-_46457233 | 0.16 |
ENSDART00000113214
|
sult1st7
|
sulfotransferase family 1, cytosolic sulfotransferase 7 |
| chr5_+_13891202 | 0.16 |
ENSDART00000193992
|
hk2
|
hexokinase 2 |
| chr6_-_17266221 | 0.16 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr6_-_16804001 | 0.16 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr6_-_17052116 | 0.15 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr1_+_12335816 | 0.15 |
ENSDART00000067086
|
nansa
|
N-acetylneuraminic acid synthase a |
| chr13_+_43650632 | 0.14 |
ENSDART00000141024
|
zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr6_-_17179637 | 0.14 |
ENSDART00000153710
|
pimr19
|
Pim proto-oncogene, serine/threonine kinase, related 19 |
| chr11_-_3308569 | 0.14 |
ENSDART00000036581
|
cdk2
|
cyclin-dependent kinase 2 |
| chr5_-_37103487 | 0.14 |
ENSDART00000149211
|
il13ra2
|
interleukin 13 receptor, alpha 2 |
| chr22_-_29204960 | 0.14 |
ENSDART00000131386
|
pvalb7
|
parvalbumin 7 |
| chr6_-_16868834 | 0.13 |
ENSDART00000156503
|
pimr41
|
Pim proto-oncogene, serine/threonine kinase, related 41 |
| chr21_+_20903244 | 0.13 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr9_+_11587629 | 0.13 |
ENSDART00000102508
ENSDART00000146585 |
si:dkey-69o16.5
|
si:dkey-69o16.5 |
| chr6_-_16948040 | 0.13 |
ENSDART00000156433
|
pimr30
|
Pim proto-oncogene, serine/threonine kinase, related 30 |
| chr7_-_17412559 | 0.12 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr13_+_24829996 | 0.12 |
ENSDART00000147194
|
si:ch211-223p8.8
|
si:ch211-223p8.8 |
| chr7_-_35083184 | 0.12 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr21_-_43171249 | 0.12 |
ENSDART00000021037
ENSDART00000148630 |
hspa4a
|
heat shock protein 4a |
| chr2_-_51511494 | 0.11 |
ENSDART00000161392
ENSDART00000160877 |
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr17_-_46817295 | 0.10 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr3_-_27065477 | 0.10 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr17_-_46933567 | 0.10 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr2_-_51611344 | 0.10 |
ENSDART00000158137
|
si:ch211-9d9.8
|
si:ch211-9d9.8 |
| chr20_+_37294112 | 0.09 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr22_+_2239254 | 0.08 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr10_+_27068251 | 0.08 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr10_+_29770120 | 0.08 |
ENSDART00000100032
ENSDART00000193205 |
hyou1
|
hypoxia up-regulated 1 |
| chr16_-_31985965 | 0.07 |
ENSDART00000161944
|
si:dkey-40m6.14
|
si:dkey-40m6.14 |
| chr24_-_20652473 | 0.07 |
ENSDART00000179984
|
nktr
|
natural killer cell triggering receptor |
| chr18_-_18524624 | 0.07 |
ENSDART00000157834
|
lonp2
|
lon peptidase 2, peroxisomal |
| chr3_+_52737565 | 0.06 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr4_+_47178124 | 0.06 |
ENSDART00000170816
|
si:dkey-26m3.2
|
si:dkey-26m3.2 |
| chr18_+_14684115 | 0.06 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr14_-_30490465 | 0.06 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr8_-_48741106 | 0.06 |
ENSDART00000164203
|
pimr180
|
Pim proto-oncogene, serine/threonine kinase, related 180 |
| chr14_+_8725216 | 0.06 |
ENSDART00000157630
|
pimr57
|
Pim proto-oncogene, serine/threonine kinase, related 57 |
| chr20_+_10727022 | 0.05 |
ENSDART00000104185
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr19_+_10860485 | 0.05 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr19_-_3783141 | 0.05 |
ENSDART00000162260
|
btr19
|
bloodthirsty-related gene family, member 19 |
| chr22_+_29204885 | 0.05 |
ENSDART00000133409
ENSDART00000059820 |
ncf4
|
neutrophil cytosolic factor 4 |
| chr11_+_44356707 | 0.04 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr22_-_16658996 | 0.04 |
ENSDART00000138429
|
si:dkey-38n4.2
|
si:dkey-38n4.2 |
| chr7_-_22699716 | 0.04 |
ENSDART00000193773
|
BX470211.5
|
|
| chr20_-_42534153 | 0.04 |
ENSDART00000061122
|
rfx6
|
regulatory factor X, 6 |
| chr10_-_4961923 | 0.03 |
ENSDART00000050177
ENSDART00000146066 |
snx30
|
sorting nexin family member 30 |
| chr24_+_35787629 | 0.03 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr23_+_21261313 | 0.03 |
ENSDART00000104268
ENSDART00000159046 |
emc1
|
ER membrane protein complex subunit 1 |
| chr15_+_15771418 | 0.03 |
ENSDART00000153831
|
si:ch211-33e4.3
|
si:ch211-33e4.3 |
| chr5_-_67911111 | 0.03 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr7_-_52388734 | 0.02 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr5_-_12824607 | 0.02 |
ENSDART00000190310
|
p2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr21_-_30408775 | 0.02 |
ENSDART00000101037
|
nhp2
|
NHP2 ribonucleoprotein homolog (yeast) |
| chr22_-_29205327 | 0.01 |
ENSDART00000183161
ENSDART00000189515 |
pvalb7
|
parvalbumin 7 |
| chr13_-_25198025 | 0.00 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.5 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.4 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.2 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.4 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.0 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.5 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.4 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.4 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.5 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.4 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0031782 | type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.2 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.5 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |