Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
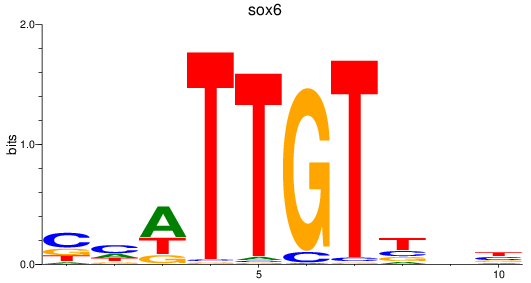
Results for sox6
Z-value: 0.98
Transcription factors associated with sox6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox6
|
ENSDARG00000015536 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox6 | dr11_v1_chr7_+_27250012_27250012 | -0.39 | 1.0e-01 | Click! |
Activity profile of sox6 motif
Sorted Z-values of sox6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22339582 | 5.26 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr9_-_22318511 | 5.09 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr9_-_22272181 | 4.83 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr5_-_30615901 | 3.79 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr1_-_55810730 | 3.68 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr10_+_44042033 | 3.56 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr7_+_39444843 | 3.38 |
ENSDART00000143999
ENSDART00000173554 ENSDART00000173698 ENSDART00000173754 ENSDART00000144075 ENSDART00000138192 ENSDART00000145457 ENSDART00000141750 ENSDART00000103056 ENSDART00000142946 ENSDART00000173748 |
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr23_+_27068225 | 2.95 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr12_-_22524388 | 2.31 |
ENSDART00000020942
|
shbg
|
sex hormone-binding globulin |
| chr2_+_3472832 | 2.31 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr15_-_576135 | 2.22 |
ENSDART00000124170
|
cbln20
|
cerebellin 20 |
| chr4_-_27301356 | 2.05 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr17_+_33226955 | 2.04 |
ENSDART00000063333
|
pomca
|
proopiomelanocortin a |
| chr21_-_41870029 | 2.04 |
ENSDART00000182035
|
endou2
|
endonuclease, polyU-specific 2 |
| chr7_+_48761875 | 1.71 |
ENSDART00000003690
|
acana
|
aggrecan a |
| chr14_-_33872092 | 1.69 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr14_-_2217285 | 1.68 |
ENSDART00000157949
ENSDART00000166150 ENSDART00000054891 ENSDART00000183268 |
pcdh2ab2
pcdh2ab2
|
protocadherin 2 alpha b2 protocadherin 2 alpha b2 |
| chr20_-_40754794 | 1.61 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr3_+_29641181 | 1.46 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr3_-_35602233 | 1.43 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr6_-_51101834 | 1.34 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr20_-_14665002 | 1.33 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr7_-_24520866 | 1.29 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr24_-_29868151 | 1.28 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr6_+_54358529 | 1.24 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr10_+_21780250 | 1.22 |
ENSDART00000183782
|
pcdh1g9
|
protocadherin 1 gamma 9 |
| chr12_-_30523216 | 1.21 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr10_+_21677058 | 1.18 |
ENSDART00000171499
ENSDART00000157516 |
pcdh1gb2
|
protocadherin 1 gamma b 2 |
| chr23_+_3538463 | 1.17 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr14_+_51056605 | 1.16 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr21_-_41028665 | 1.15 |
ENSDART00000190531
|
plac8l1
|
PLAC8-like 1 |
| chr25_-_3503164 | 1.15 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr7_+_48761646 | 1.14 |
ENSDART00000017467
|
acana
|
aggrecan a |
| chr4_-_5291256 | 1.12 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr13_-_20540790 | 1.12 |
ENSDART00000131467
|
si:ch1073-126c3.2
|
si:ch1073-126c3.2 |
| chr10_-_22831611 | 1.10 |
ENSDART00000160115
|
per1a
|
period circadian clock 1a |
| chr12_+_45677293 | 1.10 |
ENSDART00000152850
ENSDART00000153047 |
si:ch73-111m19.2
|
si:ch73-111m19.2 |
| chr12_+_40810418 | 1.10 |
ENSDART00000183393
|
CDH18
|
cadherin 18 |
| chr10_+_21722892 | 1.05 |
ENSDART00000162855
|
pcdh1g13
|
protocadherin 1 gamma 13 |
| chr15_+_17756113 | 1.03 |
ENSDART00000155197
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr1_+_41690402 | 0.98 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr10_+_20113830 | 0.96 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr4_+_18804317 | 0.94 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr13_-_11699530 | 0.93 |
ENSDART00000192161
|
slc39a8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr6_+_39812475 | 0.92 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr9_-_18742704 | 0.92 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr18_+_21122818 | 0.90 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr20_-_27864964 | 0.89 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr14_-_2318590 | 0.89 |
ENSDART00000192735
|
pcdh2ab8
|
protocadherin 2 alpha b 8 |
| chr12_-_33354409 | 0.87 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr15_+_29123031 | 0.85 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr4_-_77432218 | 0.84 |
ENSDART00000158683
|
slco1d1
|
solute carrier organic anion transporter family, member 1D1 |
| chr8_-_19649617 | 0.83 |
ENSDART00000189033
|
fam78bb
|
family with sequence similarity 78, member B b |
| chr12_+_5081759 | 0.82 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr10_-_42131408 | 0.81 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr23_-_7826849 | 0.80 |
ENSDART00000157612
|
myt1b
|
myelin transcription factor 1b |
| chr11_+_35364445 | 0.80 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr21_-_35853245 | 0.80 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr14_+_11457500 | 0.79 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr2_-_30784198 | 0.77 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr9_+_50001746 | 0.75 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr7_-_73852594 | 0.74 |
ENSDART00000183194
|
zgc:165555
|
zgc:165555 |
| chr9_-_23891102 | 0.73 |
ENSDART00000186799
|
asb18
|
ankyrin repeat and SOCS box containing 18 |
| chr25_+_13620555 | 0.71 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr15_-_5563551 | 0.67 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr24_-_24724233 | 0.67 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr11_+_14280598 | 0.66 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr7_-_6604623 | 0.65 |
ENSDART00000172874
|
kcnj10a
|
potassium inwardly-rectifying channel, subfamily J, member 10a |
| chr23_+_42813415 | 0.64 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr13_-_27916439 | 0.64 |
ENSDART00000139081
ENSDART00000087097 |
ogfrl1
|
opioid growth factor receptor-like 1 |
| chr9_-_18743012 | 0.63 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr6_-_25180438 | 0.62 |
ENSDART00000159696
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr3_+_49074008 | 0.62 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr6_-_19351495 | 0.61 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr23_-_637347 | 0.61 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr5_+_31480342 | 0.60 |
ENSDART00000098197
|
si:dkey-220k22.1
|
si:dkey-220k22.1 |
| chr22_+_1028724 | 0.60 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr10_+_5744941 | 0.60 |
ENSDART00000159769
ENSDART00000184734 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr1_-_17735861 | 0.59 |
ENSDART00000018238
|
pdlim3a
|
PDZ and LIM domain 3a |
| chr2_+_56463167 | 0.59 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr9_+_8942258 | 0.58 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr25_+_34862225 | 0.57 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr11_+_10541258 | 0.56 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr2_+_26240631 | 0.56 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr5_-_30984010 | 0.56 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr7_-_53117131 | 0.54 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr11_+_6152643 | 0.54 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr17_-_30666037 | 0.52 |
ENSDART00000156509
|
alkal2b
|
ALK and LTK ligand 2b |
| chr20_+_37295006 | 0.51 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr19_+_24575077 | 0.51 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr12_+_33151246 | 0.50 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr17_+_27158706 | 0.50 |
ENSDART00000151829
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr5_-_26181863 | 0.49 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_+_55221593 | 0.49 |
ENSDART00000073638
|
tmc2a
|
transmembrane channel-like 2a |
| chr14_+_48812189 | 0.48 |
ENSDART00000161454
|
mmp17b
|
matrix metallopeptidase 17b |
| chr18_+_11970987 | 0.47 |
ENSDART00000144111
|
si:dkeyp-2c8.3
|
si:dkeyp-2c8.3 |
| chr21_-_43550120 | 0.46 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr2_-_24269911 | 0.45 |
ENSDART00000099532
|
myh7
|
myosin heavy chain 7 |
| chr2_-_10896745 | 0.42 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr7_+_24520518 | 0.42 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr9_+_13986427 | 0.41 |
ENSDART00000147200
|
cd28
|
CD28 molecule |
| chr16_-_21799407 | 0.41 |
ENSDART00000123717
|
FP015862.4
|
|
| chr14_+_11946395 | 0.41 |
ENSDART00000193290
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr3_+_3545825 | 0.41 |
ENSDART00000109060
|
CR589947.1
|
|
| chr16_+_27349585 | 0.40 |
ENSDART00000142573
|
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr16_-_22006996 | 0.40 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr2_-_5942355 | 0.39 |
ENSDART00000110469
|
tmem125b
|
transmembrane protein 125b |
| chr11_-_9808356 | 0.39 |
ENSDART00000167465
|
nlgn1
|
neuroligin 1 |
| chr6_+_54888493 | 0.39 |
ENSDART00000113331
|
nav1b
|
neuron navigator 1b |
| chr25_+_35502552 | 0.38 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr23_+_5499307 | 0.38 |
ENSDART00000170361
|
tead3a
|
TEA domain family member 3 a |
| chr21_-_21600640 | 0.38 |
ENSDART00000079495
|
or133-1
|
odorant receptor, family H, subfamily 133, member 1 |
| chr7_+_38255418 | 0.37 |
ENSDART00000052354
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr7_-_24373662 | 0.35 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr7_-_13362590 | 0.35 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr1_-_18615063 | 0.35 |
ENSDART00000014916
|
klf3
|
Kruppel-like factor 3 (basic) |
| chr5_-_21422390 | 0.34 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr9_-_9242777 | 0.34 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr17_+_8899016 | 0.34 |
ENSDART00000150214
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr15_+_37331585 | 0.33 |
ENSDART00000170715
|
zgc:171592
|
zgc:171592 |
| chr15_+_5901970 | 0.32 |
ENSDART00000114134
|
wrb
|
tryptophan rich basic protein |
| chr22_-_15693085 | 0.32 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr13_+_33368503 | 0.31 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr23_-_5685023 | 0.31 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr12_-_31009315 | 0.30 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7 like 2 |
| chr7_+_31145386 | 0.30 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr2_-_24270062 | 0.27 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr20_-_13722514 | 0.27 |
ENSDART00000152458
|
si:ch211-122h15.4
|
si:ch211-122h15.4 |
| chr24_+_36392784 | 0.26 |
ENSDART00000134750
|
arf2b
|
ADP-ribosylation factor 2b |
| chr2_+_29257942 | 0.26 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr8_+_16990120 | 0.26 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr14_-_31060082 | 0.26 |
ENSDART00000111601
ENSDART00000161113 |
mbnl3
|
muscleblind-like splicing regulator 3 |
| chr1_-_354115 | 0.26 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr7_-_17570923 | 0.25 |
ENSDART00000188476
ENSDART00000080624 |
nitr5
|
novel immune-type receptor 5 |
| chr5_-_28149767 | 0.25 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr16_+_25245857 | 0.23 |
ENSDART00000155220
|
klhl38b
|
kelch-like family member 38b |
| chr12_+_35654749 | 0.23 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr21_+_11105007 | 0.23 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr10_-_26163989 | 0.22 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr20_+_40237441 | 0.22 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr14_+_52565660 | 0.22 |
ENSDART00000188151
|
rpl26
|
ribosomal protein L26 |
| chr1_-_56213723 | 0.21 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr7_-_33868903 | 0.21 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr20_-_33961697 | 0.20 |
ENSDART00000061765
|
selp
|
selectin P |
| chr11_+_34523132 | 0.20 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr6_-_41138854 | 0.19 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr17_+_31611854 | 0.19 |
ENSDART00000011706
|
adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr17_-_44776224 | 0.18 |
ENSDART00000156195
|
zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr22_+_3045495 | 0.18 |
ENSDART00000164061
|
LO017843.1
|
|
| chr24_+_36392040 | 0.18 |
ENSDART00000136057
|
arf2b
|
ADP-ribosylation factor 2b |
| chr16_+_47308856 | 0.18 |
ENSDART00000170103
|
col28a1b
|
collagen, type XXVIII, alpha 1b |
| chr3_+_24189804 | 0.17 |
ENSDART00000134723
|
prr15la
|
proline rich 15-like a |
| chr24_+_36392486 | 0.15 |
ENSDART00000007381
ENSDART00000135898 |
arf2b
|
ADP-ribosylation factor 2b |
| chr5_+_42912966 | 0.15 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr20_+_733510 | 0.15 |
ENSDART00000135066
ENSDART00000015558 ENSDART00000152782 |
myo6a
|
myosin VIa |
| chr23_+_30827959 | 0.15 |
ENSDART00000033429
|
npbwr2a
|
neuropeptides B/W receptor 2a |
| chr5_-_38384289 | 0.14 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr19_-_43226124 | 0.14 |
ENSDART00000168965
|
gnrhr1
|
gonadotropin releasing hormone receptor 1 |
| chr5_+_19097194 | 0.14 |
ENSDART00000131918
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr1_-_46981134 | 0.14 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr25_-_22889519 | 0.12 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr15_+_17441734 | 0.12 |
ENSDART00000153729
|
snx19b
|
sorting nexin 19b |
| chr3_-_25814097 | 0.12 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr8_-_49499457 | 0.11 |
ENSDART00000098326
|
opn7d
|
opsin 7, group member d |
| chr4_-_63848305 | 0.11 |
ENSDART00000097308
|
BX901974.1
|
|
| chr17_-_35238359 | 0.10 |
ENSDART00000005784
|
itgb1bp1
|
integrin beta 1 binding protein 1 |
| chr15_-_41503392 | 0.09 |
ENSDART00000169351
|
si:dkey-230i18.2
|
si:dkey-230i18.2 |
| chr21_+_34119759 | 0.09 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr15_-_9321921 | 0.08 |
ENSDART00000159676
ENSDART00000033411 |
trappc4
|
trafficking protein particle complex 4 |
| chr22_-_24967348 | 0.08 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr15_+_12429206 | 0.07 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr20_+_31217495 | 0.07 |
ENSDART00000020252
|
pdia6
|
protein disulfide isomerase family A, member 6 |
| chr3_+_24190207 | 0.07 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr7_-_40144686 | 0.07 |
ENSDART00000109166
|
wdr60
|
WD repeat domain 60 |
| chr12_-_7280551 | 0.06 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr12_+_38697541 | 0.06 |
ENSDART00000188648
|
dnai2b
|
dynein, axonemal, intermediate chain 2b |
| chr22_+_30631072 | 0.06 |
ENSDART00000059970
|
zmp:0000000606
|
zmp:0000000606 |
| chr7_-_40145097 | 0.06 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr5_+_7989210 | 0.06 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr25_+_13271458 | 0.06 |
ENSDART00000188307
|
AL772241.1
|
|
| chr5_-_31875645 | 0.06 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr12_+_22657925 | 0.05 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr19_+_4051695 | 0.05 |
ENSDART00000166368
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr15_+_31481939 | 0.05 |
ENSDART00000134306
|
or102-5
|
odorant receptor, family C, subfamily 102, member 5 |
| chr2_-_13268556 | 0.05 |
ENSDART00000127693
ENSDART00000079757 |
vps4b
|
vacuolar protein sorting 4b homolog B (S. cerevisiae) |
| chr3_+_3099492 | 0.05 |
ENSDART00000154156
|
si:dkey-30g5.1
|
si:dkey-30g5.1 |
| chr8_+_17069577 | 0.04 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr7_+_24114694 | 0.04 |
ENSDART00000127177
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr13_-_22771708 | 0.04 |
ENSDART00000112900
|
si:ch211-150i13.1
|
si:ch211-150i13.1 |
| chr12_+_17042754 | 0.03 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr5_+_40835601 | 0.03 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr2_+_49457449 | 0.03 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr23_+_7710721 | 0.03 |
ENSDART00000186852
ENSDART00000161193 |
kif3b
|
kinesin family member 3B |
| chr4_+_29702703 | 0.02 |
ENSDART00000167771
|
si:ch211-214c20.1
|
si:ch211-214c20.1 |
| chr21_+_349091 | 0.02 |
ENSDART00000183693
|
ZNF462
|
zinc finger protein 462 |
| chr5_+_10046643 | 0.01 |
ENSDART00000137543
ENSDART00000186917 |
gstt2
|
glutathione S-transferase theta 2 |
| chr8_+_8671229 | 0.01 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr17_+_5623514 | 0.01 |
ENSDART00000171220
ENSDART00000176083 |
CU571310.1
|
|
| chr8_+_14158021 | 0.01 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr8_+_26205471 | 0.00 |
ENSDART00000131888
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.5 | 2.9 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.3 | 2.9 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 0.9 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.3 | 0.9 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 0.8 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 18.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 3.4 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.7 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 1.4 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.7 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.1 | 0.6 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.6 | GO:0036268 | swimming(GO:0036268) |
| 0.1 | 1.3 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.5 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 2.0 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 0.5 | GO:0070376 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.1 | 0.3 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.4 | GO:0034552 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.1 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.8 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.5 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 1.1 | GO:0043153 | response to hydrogen peroxide(GO:0042542) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0043435 | neutrophil homeostasis(GO:0001780) response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.8 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 1.0 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:0090184 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 4.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.5 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.6 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 1.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 3.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.9 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.7 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 2.9 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.3 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.3 | 0.9 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.3 | 1.3 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 18.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 0.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.6 | GO:0047256 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.1 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.1 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.5 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.1 | 0.8 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.1 | 3.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.3 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 1.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 2.0 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.9 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.5 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 2.3 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.2 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.8 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | ST G ALPHA I PATHWAY | G alpha i Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 0.5 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.9 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 0.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |