Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
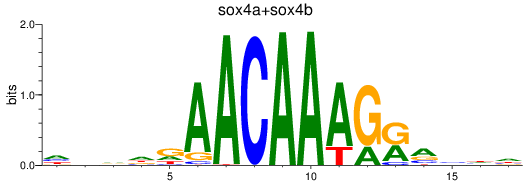
Results for sox4a+sox4b
Z-value: 0.57
Transcription factors associated with sox4a+sox4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox4a
|
ENSDARG00000004588 | SRY-box transcription factor 4a |
|
sox4b
|
ENSDARG00000098834 | SRY-box transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox4a | dr11_v1_chr19_-_28789404_28789409 | -0.52 | 2.3e-02 | Click! |
| sox4b | dr11_v1_chr16_+_68069_68124 | -0.44 | 5.7e-02 | Click! |
Activity profile of sox4a+sox4b motif
Sorted Z-values of sox4a+sox4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_3758637 | 1.32 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr9_+_15893093 | 1.16 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr23_-_3759345 | 0.92 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr6_+_46406565 | 0.81 |
ENSDART00000168440
ENSDART00000131203 ENSDART00000138567 ENSDART00000132845 |
pbrm1l
|
polybromo 1, like |
| chr19_-_46037835 | 0.81 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr8_+_17869225 | 0.77 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr1_+_17376922 | 0.76 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr5_+_43870389 | 0.72 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr19_-_18152407 | 0.71 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr5_+_44846434 | 0.71 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr2_-_24069331 | 0.68 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr13_+_48358467 | 0.63 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr8_-_16697912 | 0.62 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr3_-_27646070 | 0.61 |
ENSDART00000122031
ENSDART00000151027 |
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr16_-_41762983 | 0.60 |
ENSDART00000192936
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr7_-_55454406 | 0.57 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr8_+_29636431 | 0.57 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr14_-_2004291 | 0.56 |
ENSDART00000114039
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr4_+_15954293 | 0.56 |
ENSDART00000132695
|
si:dkey-117n7.4
|
si:dkey-117n7.4 |
| chr20_-_33790003 | 0.55 |
ENSDART00000020183
|
fam102bb
|
family with sequence similarity 102, member B, b |
| chr16_-_33104944 | 0.54 |
ENSDART00000151943
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr21_-_5077715 | 0.53 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr7_-_24995631 | 0.53 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr21_-_37027252 | 0.52 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr24_+_17142881 | 0.52 |
ENSDART00000177272
ENSDART00000192259 ENSDART00000191029 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr17_+_25849332 | 0.52 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr7_-_57332915 | 0.52 |
ENSDART00000162653
|
BX470176.1
|
|
| chr11_+_14057605 | 0.50 |
ENSDART00000166262
|
grin3bb
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3Bb |
| chr13_-_21650404 | 0.49 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr20_+_34770197 | 0.49 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr22_-_22301672 | 0.48 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr19_-_17774875 | 0.47 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr10_-_36633882 | 0.47 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr16_+_35535375 | 0.47 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr11_+_3501669 | 0.47 |
ENSDART00000160808
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr10_-_25628555 | 0.46 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr11_+_6115621 | 0.46 |
ENSDART00000165031
ENSDART00000027666 ENSDART00000161458 |
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr6_+_17789481 | 0.46 |
ENSDART00000190768
|
CR376861.1
|
|
| chr15_+_35691783 | 0.45 |
ENSDART00000183994
|
CT573366.1
|
|
| chr5_-_12093618 | 0.45 |
ENSDART00000161542
|
lrrc74b
|
leucine rich repeat containing 74B |
| chr6_-_9282080 | 0.44 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr5_-_46505691 | 0.44 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr6_+_59832786 | 0.43 |
ENSDART00000154985
ENSDART00000102148 |
ddx3b
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3b |
| chr17_+_24632440 | 0.42 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr7_+_53879191 | 0.42 |
ENSDART00000164768
|
neo1a
|
neogenin 1a |
| chr9_-_32142311 | 0.42 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr5_+_23045096 | 0.41 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr19_+_7424347 | 0.41 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr24_+_9696760 | 0.41 |
ENSDART00000140200
ENSDART00000187411 |
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr15_+_28368823 | 0.41 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr4_-_20511595 | 0.40 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr6_-_59505589 | 0.39 |
ENSDART00000170685
|
gli1
|
GLI family zinc finger 1 |
| chr2_-_20574193 | 0.39 |
ENSDART00000190448
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr19_-_17208728 | 0.39 |
ENSDART00000151228
|
stmn1a
|
stathmin 1a |
| chr21_-_2310064 | 0.39 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr12_+_48340133 | 0.39 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr13_+_28732101 | 0.39 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr23_+_36144487 | 0.38 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr13_-_5257303 | 0.38 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr11_+_30296332 | 0.38 |
ENSDART00000192843
|
ugt1b7
|
UDP glucuronosyltransferase 1 family, polypeptide B7 |
| chr17_+_7534180 | 0.38 |
ENSDART00000187512
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr22_-_10440688 | 0.38 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr19_-_7540821 | 0.38 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr5_+_44846280 | 0.38 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr13_+_2894536 | 0.36 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr14_+_6962271 | 0.36 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr22_+_12161738 | 0.36 |
ENSDART00000168857
|
ccnt2b
|
cyclin T2b |
| chr17_+_7534365 | 0.35 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr3_+_60761811 | 0.35 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr25_+_31323978 | 0.35 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr14_+_6963312 | 0.35 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr1_+_41609676 | 0.35 |
ENSDART00000183675
|
mogs
|
mannosyl-oligosaccharide glucosidase |
| chr14_-_44773864 | 0.35 |
ENSDART00000158386
|
si:dkey-109l4.3
|
si:dkey-109l4.3 |
| chr12_+_2804505 | 0.34 |
ENSDART00000152193
|
mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr21_-_9914745 | 0.34 |
ENSDART00000172124
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr17_-_4252221 | 0.34 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr4_+_48770245 | 0.34 |
ENSDART00000138053
|
znf1020
|
zinc finger protein 1020 |
| chr9_-_695000 | 0.34 |
ENSDART00000181318
ENSDART00000115030 |
dip2a
|
disco-interacting protein 2 homolog A |
| chr3_-_25054002 | 0.34 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr10_+_32058692 | 0.33 |
ENSDART00000062309
|
thap12a
|
THAP domain containing 12a |
| chr16_+_26732086 | 0.33 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr5_-_22602979 | 0.33 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr16_-_26731928 | 0.33 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr16_-_33105677 | 0.32 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr23_+_36122058 | 0.32 |
ENSDART00000184448
|
hoxc3a
|
homeobox C3a |
| chr10_-_2788668 | 0.32 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr6_+_12968101 | 0.32 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr11_+_807153 | 0.32 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr21_-_32781612 | 0.31 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr19_-_20482261 | 0.31 |
ENSDART00000056205
|
satb1a
|
SATB homeobox 1a |
| chr7_-_24875421 | 0.31 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr7_+_65876335 | 0.31 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr6_-_31224563 | 0.31 |
ENSDART00000104616
|
lepr
|
leptin receptor |
| chr21_-_2310355 | 0.31 |
ENSDART00000183326
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr22_-_7025393 | 0.30 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr3_-_21348478 | 0.30 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr6_-_12172424 | 0.30 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr5_-_22602780 | 0.30 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr17_+_44030692 | 0.30 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr22_-_9861531 | 0.29 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr8_-_51340773 | 0.29 |
ENSDART00000060633
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr23_+_25428513 | 0.29 |
ENSDART00000144554
|
fmnl3
|
formin-like 3 |
| chr15_+_28368644 | 0.29 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr1_-_43987873 | 0.29 |
ENSDART00000108821
|
CR385050.1
|
|
| chr12_-_20616160 | 0.28 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr12_+_28856151 | 0.28 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr17_+_48164536 | 0.28 |
ENSDART00000161750
ENSDART00000156923 |
plekhd1
|
pleckstrin homology domain containing, family D (with coiled-coil domains) member 1 |
| chr1_+_44173506 | 0.28 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr8_+_54135642 | 0.28 |
ENSDART00000170712
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr16_+_25342152 | 0.27 |
ENSDART00000156351
|
zfat
|
zinc finger and AT hook domain containing |
| chr19_+_43969363 | 0.26 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr25_-_35112466 | 0.26 |
ENSDART00000183481
|
CU302436.4
|
|
| chr15_+_19458982 | 0.26 |
ENSDART00000132665
|
zgc:77784
|
zgc:77784 |
| chr20_+_31269778 | 0.26 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr21_-_2341937 | 0.26 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr22_-_9860792 | 0.26 |
ENSDART00000155908
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr13_+_5978809 | 0.26 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr4_+_9177997 | 0.25 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr4_-_15003854 | 0.25 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr9_+_27329640 | 0.25 |
ENSDART00000111039
|
GTPBP8
|
si:rp71-84d19.3 |
| chr2_-_52455525 | 0.25 |
ENSDART00000160768
ENSDART00000002241 |
chico
|
chico |
| chr17_-_5860222 | 0.25 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr10_+_29849497 | 0.25 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr17_-_45378473 | 0.25 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr14_-_1958994 | 0.25 |
ENSDART00000161783
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr1_+_29766725 | 0.24 |
ENSDART00000054064
|
zc3h13
|
zinc finger CCCH-type containing 13 |
| chr4_+_9178913 | 0.24 |
ENSDART00000168558
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr25_+_28776562 | 0.24 |
ENSDART00000109702
|
slc41a2a
|
solute carrier family 41 (magnesium transporter), member 2a |
| chr21_+_4540127 | 0.24 |
ENSDART00000043431
|
nup188
|
nucleoporin 188 |
| chr4_-_178510 | 0.24 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr2_-_22966076 | 0.24 |
ENSDART00000143412
ENSDART00000146014 ENSDART00000183443 ENSDART00000191056 ENSDART00000183539 |
sap130b
|
Sin3A-associated protein b |
| chr11_+_24820542 | 0.24 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr9_+_12948511 | 0.24 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr20_-_16498991 | 0.24 |
ENSDART00000104137
|
ches1
|
checkpoint suppressor 1 |
| chr1_-_8566567 | 0.24 |
ENSDART00000114613
|
ptcd1
|
pentatricopeptide repeat domain 1 |
| chr16_+_25068576 | 0.23 |
ENSDART00000125838
|
im:7147486
|
im:7147486 |
| chr21_+_26748141 | 0.23 |
ENSDART00000169025
|
pcxa
|
pyruvate carboxylase a |
| chr16_+_35535171 | 0.23 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr10_-_1827225 | 0.23 |
ENSDART00000058627
|
epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr13_+_39188737 | 0.23 |
ENSDART00000083641
|
fam135a
|
family with sequence similarity 135, member A |
| chr16_+_6750756 | 0.22 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr4_+_13953537 | 0.22 |
ENSDART00000133596
|
pphln1
|
periphilin 1 |
| chr1_-_34335752 | 0.22 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr14_-_1955257 | 0.22 |
ENSDART00000193254
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr5_+_41477954 | 0.22 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr10_-_25591194 | 0.22 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr1_-_51720633 | 0.22 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr24_+_18714212 | 0.22 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr11_+_3959495 | 0.22 |
ENSDART00000122953
|
gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr22_-_22337382 | 0.22 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr14_+_39258569 | 0.21 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr19_+_40026959 | 0.21 |
ENSDART00000123647
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr12_+_10115964 | 0.21 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr3_-_52661242 | 0.21 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr8_-_16712111 | 0.21 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr1_+_14073891 | 0.21 |
ENSDART00000021693
|
ank2a
|
ankyrin 2a, neuronal |
| chr20_-_49681850 | 0.21 |
ENSDART00000025926
|
col12a1b
|
collagen, type XII, alpha 1b |
| chr3_-_29891218 | 0.21 |
ENSDART00000142118
|
slc25a39
|
solute carrier family 25, member 39 |
| chr5_-_32383475 | 0.21 |
ENSDART00000141294
|
fbxw2
|
F-box and WD repeat domain containing 2 |
| chr14_-_31151148 | 0.21 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr2_-_57264262 | 0.21 |
ENSDART00000183815
ENSDART00000149829 ENSDART00000088508 ENSDART00000149508 |
mbd3a
|
methyl-CpG binding domain protein 3a |
| chr2_+_35733335 | 0.21 |
ENSDART00000113489
|
rasal2
|
RAS protein activator like 2 |
| chr19_+_7636941 | 0.21 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr12_-_28848015 | 0.20 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr7_+_73827805 | 0.20 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr6_+_7322587 | 0.20 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr15_+_22435460 | 0.20 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr1_+_44173245 | 0.20 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr10_+_29849977 | 0.20 |
ENSDART00000180242
|
hspa8
|
heat shock protein 8 |
| chr7_+_20019125 | 0.20 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr7_-_69636502 | 0.20 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr5_+_20823409 | 0.20 |
ENSDART00000093185
ENSDART00000142894 |
limk2
|
LIM domain kinase 2 |
| chr8_+_4803906 | 0.19 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr2_-_3038904 | 0.19 |
ENSDART00000186795
|
guk1a
|
guanylate kinase 1a |
| chr20_+_3277620 | 0.19 |
ENSDART00000067397
ENSDART00000135194 |
ndufaf7
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 7 |
| chr14_-_41272034 | 0.19 |
ENSDART00000191709
ENSDART00000074438 |
cenpi
|
centromere protein I |
| chr8_-_51340931 | 0.19 |
ENSDART00000178209
|
kansl3
|
KAT8 regulatory NSL complex subunit 3 |
| chr6_-_19023468 | 0.19 |
ENSDART00000184729
|
sept9b
|
septin 9b |
| chr2_-_7696287 | 0.18 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr12_+_13348918 | 0.18 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr23_-_7594723 | 0.18 |
ENSDART00000115298
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr19_-_30800004 | 0.18 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr23_-_33944597 | 0.18 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr12_-_5188413 | 0.18 |
ENSDART00000161988
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr4_+_34860183 | 0.17 |
ENSDART00000123281
ENSDART00000192960 ENSDART00000140637 ENSDART00000192576 ENSDART00000188148 |
znf985
|
zinc finger protein 985 |
| chr7_-_41512999 | 0.17 |
ENSDART00000173577
|
si:dkey-10f23.2
|
si:dkey-10f23.2 |
| chr14_-_28430505 | 0.17 |
ENSDART00000192373
|
nek12
|
NIMA-related kinase 12 |
| chr16_+_26747766 | 0.17 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr15_+_34963316 | 0.17 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr8_-_20862443 | 0.17 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr4_-_20135406 | 0.17 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr5_+_1965296 | 0.17 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr5_+_36900157 | 0.16 |
ENSDART00000183533
ENSDART00000051184 |
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr4_-_26035770 | 0.16 |
ENSDART00000124514
|
usp44
|
ubiquitin specific peptidase 44 |
| chr18_+_8231138 | 0.16 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr3_-_3703572 | 0.16 |
ENSDART00000111017
|
si:ch211-163m16.7
|
si:ch211-163m16.7 |
| chr1_-_47071979 | 0.16 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr1_+_5485799 | 0.16 |
ENSDART00000022307
|
atic
|
5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase |
| chr2_+_26179096 | 0.16 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr3_+_52999962 | 0.16 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox4a+sox4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.2 | 0.8 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.7 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.4 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.4 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.4 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.1 | 0.5 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.5 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.4 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.4 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 0.8 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.3 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.3 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.5 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.3 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.4 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.2 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 0.0 | 0.3 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.2 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.2 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0046959 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.1 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0000459 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.7 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 1.4 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.2 | GO:0070570 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 2.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.1 | GO:0021694 | cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.2 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.2 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.5 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.2 | 0.5 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.5 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.4 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.1 | 0.4 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 0.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.2 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 0.5 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.3 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.4 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 1.1 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.4 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 3.0 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0032404 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 0.8 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.2 | 0.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 0.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.3 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.1 | 0.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.1 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.6 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 5.3 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 1.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |