Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
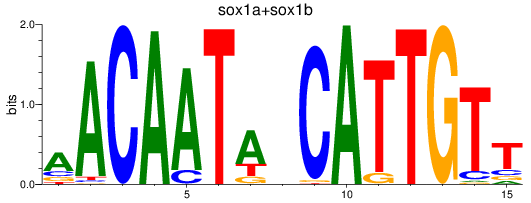
Results for sox1a+sox1b
Z-value: 0.75
Transcription factors associated with sox1a+sox1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox1b
|
ENSDARG00000008131 | SRY-box transcription factor 1b |
|
sox1a
|
ENSDARG00000069866 | SRY-box transcription factor 1a |
|
sox1a
|
ENSDARG00000110682 | SRY-box transcription factor 1a |
|
sox1a
|
ENSDARG00000114571 | SRY-box transcription factor 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox1a | dr11_v1_chr9_+_21722733_21722733 | 0.65 | 2.8e-03 | Click! |
| sox1b | dr11_v1_chr1_+_46194333_46194333 | 0.53 | 2.0e-02 | Click! |
Activity profile of sox1a+sox1b motif
Sorted Z-values of sox1a+sox1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_15440841 | 3.97 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr25_+_18475032 | 2.51 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr14_+_15768942 | 2.20 |
ENSDART00000158998
|
ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr19_-_41472228 | 2.13 |
ENSDART00000113388
|
dlx5a
|
distal-less homeobox 5a |
| chr25_+_418932 | 1.81 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr14_+_9421510 | 1.71 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr19_+_20177887 | 1.65 |
ENSDART00000008595
|
tra2a
|
transformer 2 alpha homolog |
| chr5_+_30520249 | 1.57 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr18_-_45761868 | 1.50 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr24_-_9291857 | 1.44 |
ENSDART00000149875
|
tgif1
|
TGFB-induced factor homeobox 1 |
| chr5_+_51833132 | 1.33 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr4_+_29206813 | 1.30 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr2_+_31308587 | 1.30 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr23_+_31815423 | 1.30 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr2_+_36620011 | 1.24 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr2_-_31833347 | 1.18 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr1_+_51479128 | 1.12 |
ENSDART00000028018
|
meis1a
|
Meis homeobox 1 a |
| chr6_-_37513133 | 1.09 |
ENSDART00000147826
|
jade3
|
jade family PHD finger 3 |
| chr22_+_10010292 | 1.07 |
ENSDART00000180096
|
BX324216.4
|
|
| chr24_+_21092156 | 1.07 |
ENSDART00000028542
|
ccdc191
|
coiled-coil domain containing 191 |
| chr1_+_59328030 | 0.99 |
ENSDART00000172464
|
CABZ01052576.1
|
|
| chr18_-_23875219 | 0.94 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_+_1881334 | 0.91 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr10_-_34889053 | 0.89 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr14_-_17072736 | 0.83 |
ENSDART00000106333
|
phox2bb
|
paired-like homeobox 2bb |
| chr9_+_2333927 | 0.81 |
ENSDART00000123340
|
atp5mc3a
|
ATP synthase membrane subunit c locus 3a |
| chr5_+_51833305 | 0.81 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr6_-_22146258 | 0.79 |
ENSDART00000181700
|
gga1
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 1 |
| chr7_+_50796422 | 0.78 |
ENSDART00000123868
|
dthd1
|
death domain containing 1 |
| chr14_-_9420911 | 0.77 |
ENSDART00000081158
|
sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr7_-_18877109 | 0.76 |
ENSDART00000113593
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr20_+_37300699 | 0.75 |
ENSDART00000067053
|
vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr17_-_19463355 | 0.75 |
ENSDART00000045881
|
dicer1
|
dicer 1, ribonuclease type III |
| chr11_+_37612214 | 0.74 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr7_+_61480296 | 0.74 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr18_-_23874929 | 0.73 |
ENSDART00000134910
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr23_-_24247672 | 0.71 |
ENSDART00000141552
|
zbtb17
|
zinc finger and BTB domain containing 17 |
| chr18_-_81280 | 0.70 |
ENSDART00000149810
ENSDART00000149321 |
hdc
|
histidine decarboxylase |
| chr11_+_31324335 | 0.69 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr3_+_9588705 | 0.66 |
ENSDART00000172543
ENSDART00000104875 |
trap1
|
TNF receptor-associated protein 1 |
| chr15_-_35410860 | 0.64 |
ENSDART00000191267
|
mecom
|
MDS1 and EVI1 complex locus |
| chr16_-_30826712 | 0.64 |
ENSDART00000122474
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr8_+_49096486 | 0.63 |
ENSDART00000192998
|
zgc:56525
|
zgc:56525 |
| chr20_+_37300296 | 0.58 |
ENSDART00000180789
|
vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr16_-_21047872 | 0.56 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr7_-_71331499 | 0.53 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr18_-_6766354 | 0.51 |
ENSDART00000132611
|
adm2b
|
adrenomedullin 2b |
| chr20_+_27093042 | 0.48 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr7_+_1383027 | 0.47 |
ENSDART00000143762
|
khnyn
|
KH and NYN domain containing |
| chr9_+_25853052 | 0.47 |
ENSDART00000127135
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr7_+_31425976 | 0.46 |
ENSDART00000075400
|
eif3jb
|
eukaryotic translation initiation factor 3, subunit Jb |
| chr23_-_6982966 | 0.43 |
ENSDART00000033774
ENSDART00000169965 |
edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr13_+_15581270 | 0.41 |
ENSDART00000189880
ENSDART00000190067 ENSDART00000041293 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr21_-_19915757 | 0.37 |
ENSDART00000164317
|
eri1
|
exoribonuclease 1 |
| chr15_+_24005289 | 0.36 |
ENSDART00000088648
|
myo18ab
|
myosin XVIIIAb |
| chr2_+_7715810 | 0.35 |
ENSDART00000130781
|
eif4a2
|
eukaryotic translation initiation factor 4A, isoform 2 |
| chr15_-_2493771 | 0.31 |
ENSDART00000184906
|
neu4
|
sialidase 4 |
| chr21_-_19916021 | 0.27 |
ENSDART00000065671
|
eri1
|
exoribonuclease 1 |
| chr22_+_35131890 | 0.24 |
ENSDART00000003303
ENSDART00000130581 |
rnf13
|
ring finger protein 13 |
| chr9_+_22359919 | 0.23 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr4_-_9664995 | 0.22 |
ENSDART00000180105
ENSDART00000029084 |
dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr11_+_16216909 | 0.21 |
ENSDART00000081035
ENSDART00000147190 |
slc25a26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr16_-_8927425 | 0.21 |
ENSDART00000000382
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr22_+_9994833 | 0.20 |
ENSDART00000172696
ENSDART00000192101 |
BX324216.1
|
|
| chr1_-_9134045 | 0.20 |
ENSDART00000142132
|
palb2
|
partner and localizer of BRCA2 |
| chr2_-_28420415 | 0.18 |
ENSDART00000183857
|
CABZ01056051.1
|
|
| chr20_-_40370736 | 0.14 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr11_+_37612672 | 0.11 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr19_+_14109348 | 0.06 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr20_-_46249185 | 0.06 |
ENSDART00000100544
|
taar10
|
trace amine-associated receptor 10 |
| chr7_+_57677120 | 0.05 |
ENSDART00000110623
|
arsj
|
arylsulfatase family, member J |
| chr7_-_71331690 | 0.04 |
ENSDART00000149682
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox1a+sox1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.3 | 2.1 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.3 | 1.6 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 0.7 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.2 | 0.7 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 1.3 | GO:0045658 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.2 | 0.6 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.2 | 0.8 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.2 | 1.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 1.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.9 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.1 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 2.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.8 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.3 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.8 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.5 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 0.7 | GO:0090305 | nucleic acid phosphodiester bond hydrolysis(GO:0090305) |
| 0.0 | 1.2 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 1.1 | GO:0009880 | embryonic pattern specification(GO:0009880) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.3 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 3.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 3.1 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.2 | 1.7 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.2 | 0.7 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) pre-miRNA binding(GO:0070883) |
| 0.1 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.7 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 2.1 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 1.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.7 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |