Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
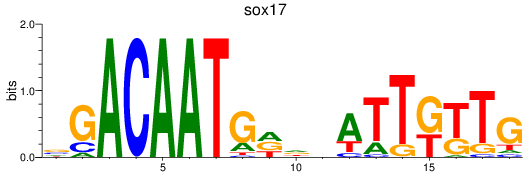
Results for sox17
Z-value: 0.78
Transcription factors associated with sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox17
|
ENSDARG00000101717 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox17 | dr11_v1_chr7_-_58776400_58776400 | 0.77 | 1.0e-04 | Click! |
Activity profile of sox17 motif
Sorted Z-values of sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22213297 | 1.79 |
ENSDART00000110656
ENSDART00000133149 |
crygm2d20
|
crystallin, gamma M2d20 |
| chr24_-_14712427 | 1.76 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr9_-_22240052 | 1.72 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr18_+_1837668 | 1.51 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr9_-_22299412 | 1.42 |
ENSDART00000139101
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr21_+_11385031 | 1.40 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr5_+_1278092 | 1.30 |
ENSDART00000147972
ENSDART00000159783 |
dnm1a
|
dynamin 1a |
| chr7_+_26100024 | 1.23 |
ENSDART00000173726
|
si:ch211-196f2.3
|
si:ch211-196f2.3 |
| chr5_+_67812062 | 1.20 |
ENSDART00000158611
|
zgc:175280
|
zgc:175280 |
| chr2_+_45636034 | 1.18 |
ENSDART00000142230
|
fndc7rs3
|
fibronectin type III domain containing 7, related sequence 3 |
| chr15_-_33896159 | 1.16 |
ENSDART00000159791
|
mag
|
myelin associated glycoprotein |
| chr2_+_55199721 | 1.16 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr16_-_28658341 | 1.16 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr16_+_43152727 | 1.11 |
ENSDART00000125590
ENSDART00000154493 |
adam22
|
ADAM metallopeptidase domain 22 |
| chr25_+_4760489 | 1.10 |
ENSDART00000167399
|
FP245455.1
|
|
| chr7_+_23495986 | 1.08 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr9_+_34641237 | 1.07 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr3_-_31079186 | 1.04 |
ENSDART00000145636
ENSDART00000140569 |
ELOB (1 of many)
elob
|
elongin B elongin B |
| chr16_+_5184402 | 1.04 |
ENSDART00000156685
|
soga3a
|
SOGA family member 3a |
| chr10_-_2522588 | 1.00 |
ENSDART00000081926
|
CU856539.1
|
|
| chr7_+_21841037 | 0.99 |
ENSDART00000077503
|
tm4sf5
|
transmembrane 4 L six family member 5 |
| chr18_+_10926197 | 0.93 |
ENSDART00000192387
|
ttc38
|
tetratricopeptide repeat domain 38 |
| chr3_+_18097700 | 0.89 |
ENSDART00000021634
|
wfikkn2a
|
info WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2a |
| chr22_+_14051245 | 0.87 |
ENSDART00000043711
ENSDART00000164259 |
aox6
|
aldehyde oxidase 6 |
| chr5_-_69004007 | 0.86 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr15_+_19652807 | 0.84 |
ENSDART00000134321
ENSDART00000054426 |
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr21_+_6328801 | 0.82 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr7_+_17992455 | 0.81 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr5_-_20678300 | 0.80 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr10_-_22845485 | 0.80 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr22_+_14051894 | 0.80 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr12_-_7806007 | 0.79 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr17_+_25289431 | 0.79 |
ENSDART00000161002
|
kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr4_-_50301491 | 0.78 |
ENSDART00000163146
|
si:ch211-197e7.3
|
si:ch211-197e7.3 |
| chr10_-_3258073 | 0.77 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr12_+_2870671 | 0.76 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr5_-_32274383 | 0.76 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr17_+_51499789 | 0.75 |
ENSDART00000187701
|
CABZ01067581.1
|
|
| chr9_-_22069364 | 0.74 |
ENSDART00000101938
|
crygm2b
|
crystallin, gamma M2b |
| chr18_-_15771551 | 0.74 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr11_-_44979281 | 0.74 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr17_-_6519423 | 0.73 |
ENSDART00000175626
|
CR628323.3
|
|
| chr3_-_34084387 | 0.72 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr14_+_36738069 | 0.71 |
ENSDART00000105590
|
tdo2a
|
tryptophan 2,3-dioxygenase a |
| chr17_-_15382704 | 0.70 |
ENSDART00000005313
|
zgc:85722
|
zgc:85722 |
| chr5_-_32273777 | 0.70 |
ENSDART00000181242
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr22_-_6884981 | 0.70 |
ENSDART00000124219
|
FO904898.4
|
|
| chr9_-_32813177 | 0.68 |
ENSDART00000012694
|
mxc
|
myxovirus (influenza virus) resistance C |
| chr11_-_40742424 | 0.67 |
ENSDART00000173399
ENSDART00000021369 |
tas1r3
|
taste receptor, type 1, member 3 |
| chr16_-_25563508 | 0.67 |
ENSDART00000189642
|
atxn1b
|
ataxin 1b |
| chr7_+_49865049 | 0.67 |
ENSDART00000164165
|
slc1a2a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2a |
| chr7_+_17992640 | 0.66 |
ENSDART00000173679
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr11_-_472547 | 0.65 |
ENSDART00000005923
|
zgc:77375
|
zgc:77375 |
| chr4_-_14328997 | 0.65 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr1_+_37391141 | 0.64 |
ENSDART00000083593
ENSDART00000168647 |
sparcl1
|
SPARC-like 1 |
| chr18_+_45504362 | 0.64 |
ENSDART00000140089
|
cngb1a
|
cyclic nucleotide gated channel beta 1a |
| chr1_-_44413629 | 0.64 |
ENSDART00000192747
|
CT583626.1
|
|
| chr5_-_68022631 | 0.64 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr16_-_50175069 | 0.63 |
ENSDART00000192979
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr14_-_3174570 | 0.62 |
ENSDART00000163585
|
csf1ra
|
colony stimulating factor 1 receptor, a |
| chr8_+_6967108 | 0.62 |
ENSDART00000004588
|
asic1a
|
acid-sensing (proton-gated) ion channel 1a |
| chr11_+_6152643 | 0.61 |
ENSDART00000012789
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr17_-_33289304 | 0.61 |
ENSDART00000135118
ENSDART00000040346 |
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr9_-_49464993 | 0.61 |
ENSDART00000181471
|
xirp2b
|
xin actin binding repeat containing 2b |
| chr19_-_15397946 | 0.61 |
ENSDART00000143480
|
hivep3a
|
human immunodeficiency virus type I enhancer binding protein 3a |
| chr11_-_29833698 | 0.60 |
ENSDART00000079149
|
xk
|
X-linked Kx blood group (McLeod syndrome) |
| chr1_+_44800298 | 0.59 |
ENSDART00000073712
ENSDART00000187054 |
tmem187
|
transmembrane protein 187 |
| chr15_-_33904831 | 0.59 |
ENSDART00000164333
ENSDART00000165404 |
mag
|
myelin associated glycoprotein |
| chr11_-_45385803 | 0.59 |
ENSDART00000173329
|
trappc10
|
trafficking protein particle complex 10 |
| chr2_-_12502495 | 0.58 |
ENSDART00000186781
|
dpp6b
|
dipeptidyl-peptidase 6b |
| chr9_-_22057658 | 0.57 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr21_+_11105007 | 0.57 |
ENSDART00000128859
ENSDART00000193105 |
prlra
|
prolactin receptor a |
| chr20_+_46867816 | 0.57 |
ENSDART00000161469
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr19_-_36234185 | 0.56 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr9_-_16218097 | 0.55 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr6_+_30091811 | 0.54 |
ENSDART00000088403
|
meltf
|
melanotransferrin |
| chr1_+_53321878 | 0.54 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr6_+_40714811 | 0.54 |
ENSDART00000153868
|
ccdc36
|
coiled-coil domain containing 36 |
| chr21_+_18353703 | 0.54 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr5_-_26181863 | 0.54 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr9_-_46276626 | 0.53 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr13_+_29462249 | 0.53 |
ENSDART00000147903
|
lrit1a
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1a |
| chr6_+_37894914 | 0.53 |
ENSDART00000148817
|
oca2
|
oculocutaneous albinism II |
| chr6_-_442163 | 0.53 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr12_-_41662327 | 0.53 |
ENSDART00000191602
ENSDART00000170423 |
jakmip3
|
Janus kinase and microtubule interacting protein 3 |
| chr21_-_7178348 | 0.52 |
ENSDART00000187467
|
fam69b
|
family with sequence similarity 69, member B |
| chr11_+_11201096 | 0.52 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr3_+_15279016 | 0.52 |
ENSDART00000182005
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr6_-_7842078 | 0.52 |
ENSDART00000065507
|
plppr2b
|
phospholipid phosphatase related 2b |
| chr17_-_52595932 | 0.51 |
ENSDART00000127225
|
si:ch211-173a9.7
|
si:ch211-173a9.7 |
| chr19_-_657439 | 0.51 |
ENSDART00000167100
|
slc6a18
|
solute carrier family 6 (neutral amino acid transporter), member 18 |
| chr7_-_24373662 | 0.50 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr25_-_1720736 | 0.50 |
ENSDART00000097256
|
SLC6A13
|
solute carrier family 6 member 13 |
| chr15_+_863931 | 0.50 |
ENSDART00000154083
|
si:dkey-7i4.12
|
si:dkey-7i4.12 |
| chr3_+_36366053 | 0.50 |
ENSDART00000181140
|
rgs9a
|
regulator of G protein signaling 9a |
| chr7_-_39378903 | 0.49 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr1_+_6817292 | 0.49 |
ENSDART00000145822
ENSDART00000092118 |
erbb4a
|
erb-b2 receptor tyrosine kinase 4a |
| chr15_-_37543591 | 0.49 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr2_+_22409249 | 0.49 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr9_+_34127005 | 0.48 |
ENSDART00000167384
ENSDART00000078065 |
f5
|
coagulation factor V |
| chr4_-_3064101 | 0.48 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr15_-_43768776 | 0.47 |
ENSDART00000170398
|
grm5b
|
glutamate receptor, metabotropic 5b |
| chr17_+_50127648 | 0.46 |
ENSDART00000156460
|
galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr1_+_37391716 | 0.46 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr1_-_23110740 | 0.46 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr23_+_25201077 | 0.45 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr10_+_15454745 | 0.45 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr1_-_46981134 | 0.45 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr5_-_69314495 | 0.45 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr7_-_3927813 | 0.45 |
ENSDART00000172991
ENSDART00000173392 |
si:dkey-88n24.12
|
si:dkey-88n24.12 |
| chr11_-_575786 | 0.45 |
ENSDART00000019997
|
mkrn2
|
makorin, ring finger protein, 2 |
| chr7_+_66565572 | 0.44 |
ENSDART00000180250
|
tmem176l.3b
|
transmembrane protein 176l.3b |
| chr22_-_26175237 | 0.44 |
ENSDART00000108737
|
c3b.2
|
complement component c3b, tandem duplicate 2 |
| chr24_-_38079261 | 0.44 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr23_+_34321237 | 0.44 |
ENSDART00000173272
|
plxna1a
|
plexin A1a |
| chr25_+_20140926 | 0.44 |
ENSDART00000121989
|
cald1b
|
caldesmon 1b |
| chr5_-_33606729 | 0.43 |
ENSDART00000137073
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr18_-_16181952 | 0.43 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr16_+_11623956 | 0.43 |
ENSDART00000137788
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr4_-_49582758 | 0.43 |
ENSDART00000180834
ENSDART00000187608 |
si:dkey-159n16.2
|
si:dkey-159n16.2 |
| chr12_-_14551077 | 0.43 |
ENSDART00000188717
|
FO704673.1
|
|
| chr17_-_6399920 | 0.43 |
ENSDART00000022010
|
hivep2b
|
human immunodeficiency virus type I enhancer binding protein 2b |
| chr22_+_1868983 | 0.42 |
ENSDART00000160135
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr11_+_575665 | 0.42 |
ENSDART00000122133
|
mkrn2os.1
|
MKRN2 opposite strand, tandem duplicate 1 |
| chr20_+_18945057 | 0.41 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr21_-_21530868 | 0.41 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr7_-_52096498 | 0.41 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr6_-_39158953 | 0.40 |
ENSDART00000050682
|
stat2
|
signal transducer and activator of transcription 2 |
| chr15_+_16521785 | 0.40 |
ENSDART00000062191
|
galnt17
|
polypeptide N-acetylgalactosaminyltransferase 17 |
| chr24_-_33276139 | 0.40 |
ENSDART00000128943
|
nrbp2b
|
nuclear receptor binding protein 2b |
| chr3_+_15773991 | 0.39 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr19_-_22498629 | 0.39 |
ENSDART00000185370
|
pleca
|
plectin a |
| chr7_-_68373495 | 0.39 |
ENSDART00000167440
|
zfhx3
|
zinc finger homeobox 3 |
| chr3_-_12381271 | 0.39 |
ENSDART00000171068
ENSDART00000157475 |
coro7
|
coronin 7 |
| chr11_-_45385602 | 0.38 |
ENSDART00000166691
|
trappc10
|
trafficking protein particle complex 10 |
| chr24_+_26406770 | 0.38 |
ENSDART00000110011
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr8_-_9684872 | 0.38 |
ENSDART00000132158
|
ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr24_-_25673405 | 0.38 |
ENSDART00000186081
ENSDART00000110241 ENSDART00000142351 |
cnksr2a
|
connector enhancer of kinase suppressor of Ras 2a |
| chr14_+_29609245 | 0.38 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr6_+_54538948 | 0.38 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr21_-_1742159 | 0.37 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr21_-_39628771 | 0.37 |
ENSDART00000183995
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr2_-_9260002 | 0.37 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr2_-_51630555 | 0.37 |
ENSDART00000171746
|
pigr
|
polymeric immunoglobulin receptor |
| chr17_-_52594756 | 0.37 |
ENSDART00000167288
|
si:ch211-173a9.7
|
si:ch211-173a9.7 |
| chr1_-_656693 | 0.37 |
ENSDART00000170483
ENSDART00000166786 |
appa
|
amyloid beta (A4) precursor protein a |
| chr19_-_22621991 | 0.36 |
ENSDART00000175106
|
pleca
|
plectin a |
| chr11_-_42134968 | 0.36 |
ENSDART00000187115
|
FP325130.1
|
|
| chr16_-_52736549 | 0.36 |
ENSDART00000146607
|
azin1a
|
antizyme inhibitor 1a |
| chr14_+_25464681 | 0.36 |
ENSDART00000067500
ENSDART00000187601 |
si:dkey-280e21.3
|
si:dkey-280e21.3 |
| chr3_-_8510201 | 0.36 |
ENSDART00000009151
|
CABZ01064671.1
|
|
| chr19_+_10493223 | 0.36 |
ENSDART00000180826
|
si:ch211-171h4.6
|
si:ch211-171h4.6 |
| chr17_+_13031497 | 0.35 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr4_-_75803810 | 0.35 |
ENSDART00000163517
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr22_+_12361317 | 0.35 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr14_-_1313480 | 0.35 |
ENSDART00000097748
|
il21
|
interleukin 21 |
| chr5_-_30074332 | 0.35 |
ENSDART00000147963
|
bco2a
|
beta-carotene oxygenase 2a |
| chr19_+_9305964 | 0.35 |
ENSDART00000136241
|
si:ch73-15n24.1
|
si:ch73-15n24.1 |
| chr14_+_2076215 | 0.34 |
ENSDART00000190062
|
CT573264.1
|
|
| chr23_-_45319592 | 0.34 |
ENSDART00000189861
|
ccdc171
|
coiled-coil domain containing 171 |
| chr5_+_45990046 | 0.34 |
ENSDART00000084024
|
sv2c
|
synaptic vesicle glycoprotein 2C |
| chr1_-_46650022 | 0.34 |
ENSDART00000148759
ENSDART00000053222 |
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr23_-_10048533 | 0.34 |
ENSDART00000166663
|
plxnb1a
|
plexin b1a |
| chr4_-_5371639 | 0.34 |
ENSDART00000150697
|
si:dkey-14d8.1
|
si:dkey-14d8.1 |
| chr19_-_41213718 | 0.34 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr1_-_44037843 | 0.34 |
ENSDART00000160276
|
si:ch73-109d9.3
|
si:ch73-109d9.3 |
| chr11_-_8320868 | 0.33 |
ENSDART00000160570
|
si:ch211-247n2.1
|
si:ch211-247n2.1 |
| chr2_-_17492486 | 0.33 |
ENSDART00000189464
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr5_-_5243079 | 0.33 |
ENSDART00000130576
ENSDART00000164377 |
mvb12ba
|
multivesicular body subunit 12Ba |
| chr4_-_38476901 | 0.33 |
ENSDART00000167124
|
si:ch211-209n20.59
|
si:ch211-209n20.59 |
| chr17_-_19963718 | 0.33 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr21_+_24536233 | 0.33 |
ENSDART00000145448
ENSDART00000109886 ENSDART00000135191 |
anapc13
|
anaphase promoting complex subunit 13 |
| chr23_+_16774589 | 0.32 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr4_-_2975461 | 0.32 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr5_-_8602937 | 0.32 |
ENSDART00000157765
|
rictora
|
RPTOR independent companion of MTOR, complex 2 a |
| chr25_+_22682065 | 0.32 |
ENSDART00000129484
|
ush1c
|
Usher syndrome 1C |
| chr2_+_36007449 | 0.32 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr20_-_45062514 | 0.32 |
ENSDART00000183529
ENSDART00000182955 |
klhl29
|
kelch-like family member 29 |
| chr21_-_2248239 | 0.32 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr11_-_25213651 | 0.32 |
ENSDART00000097316
ENSDART00000152186 |
myh7ba
|
myosin, heavy chain 7B, cardiac muscle, beta a |
| chr7_+_18017756 | 0.32 |
ENSDART00000173717
|
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr12_-_31103906 | 0.31 |
ENSDART00000189099
ENSDART00000121527 ENSDART00000185635 ENSDART00000183754 |
tcf7l2
|
transcription factor 7 like 2 |
| chr22_-_26236188 | 0.31 |
ENSDART00000162640
ENSDART00000167169 ENSDART00000138595 |
c3b.1
|
complement component c3b, tandem duplicate 1 |
| chr1_+_29281764 | 0.31 |
ENSDART00000112106
|
fam155a
|
family with sequence similarity 155, member A |
| chr20_-_35040041 | 0.31 |
ENSDART00000131919
|
kif26bb
|
kinesin family member 26Bb |
| chr17_-_20558961 | 0.31 |
ENSDART00000155993
|
sh3pxd2ab
|
SH3 and PX domains 2Ab |
| chr11_-_42918971 | 0.30 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr1_-_12278056 | 0.30 |
ENSDART00000139336
ENSDART00000137463 |
cplx2l
|
complexin 2, like |
| chr23_-_27101600 | 0.30 |
ENSDART00000139231
|
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr22_+_1352192 | 0.29 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr17_+_51520073 | 0.28 |
ENSDART00000189646
ENSDART00000170951 ENSDART00000189492 |
pxdn
|
peroxidasin |
| chr23_-_26652273 | 0.28 |
ENSDART00000112124
ENSDART00000111029 |
hspg2
|
heparan sulfate proteoglycan 2 |
| chr2_+_51818039 | 0.28 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr4_+_25950372 | 0.28 |
ENSDART00000125767
|
metap2a
|
methionyl aminopeptidase 2a |
| chr4_-_24031924 | 0.28 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr22_+_7923713 | 0.28 |
ENSDART00000167210
|
CABZ01034691.1
|
|
| chr6_+_4299164 | 0.27 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr21_-_39546737 | 0.27 |
ENSDART00000006971
|
sept4a
|
septin 4a |
| chr8_-_45277370 | 0.27 |
ENSDART00000146364
|
adamts13
|
ADAM metallopeptidase with thrombospondin type 1 motif, 13 |
| chr22_+_12361489 | 0.27 |
ENSDART00000182483
|
r3hdm1
|
R3H domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:1903826 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.2 | 1.0 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 0.5 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 0.6 | GO:0042481 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) regulation of odontogenesis(GO:0042481) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) |
| 0.1 | 0.6 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 1.4 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 1.7 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 0.4 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.3 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.7 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.1 | 0.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 0.3 | GO:0044803 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 1.6 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.1 | 0.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.2 | GO:0060907 | macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) |
| 0.1 | 0.2 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.3 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.1 | 0.5 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.1 | 0.4 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.5 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.1 | 0.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.2 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.7 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.3 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.3 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 5.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.8 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.3 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 1.2 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.0 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.4 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.1 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.5 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.5 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.1 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.0 | GO:0006368 | transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.1 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.6 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.4 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 1.0 | GO:0048741 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.0 | 0.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.0 | 0.3 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.7 | GO:0043534 | blood vessel endothelial cell migration(GO:0043534) |
| 0.0 | 0.3 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.0 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0070831 | basement membrane assembly(GO:0070831) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.2 | 0.7 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.2 | 1.0 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.2 | 1.0 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 1.7 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.6 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0032997 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.0 | 0.8 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.6 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.8 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 1.2 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 0.1 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.2 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.5 | 1.4 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 1.2 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 0.7 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 1.7 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 0.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.6 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.9 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.8 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 7.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.4 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.5 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.2 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.7 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0019767 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 0.0 | 0.3 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.3 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.5 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.8 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.4 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.5 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.9 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 0.6 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |