Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
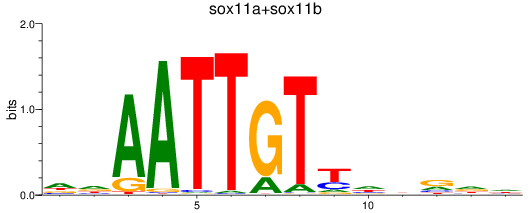
Results for sox11a+sox11b
Z-value: 1.00
Transcription factors associated with sox11a+sox11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox11a
|
ENSDARG00000077811 | SRY-box transcription factor 11a |
|
sox11b
|
ENSDARG00000095743 | SRY-box transcription factor 11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox11b | dr11_v1_chr20_-_30035326_30035326 | -0.42 | 7.7e-02 | Click! |
| sox11a | dr11_v1_chr17_-_35881841_35881841 | -0.39 | 9.7e-02 | Click! |
Activity profile of sox11a+sox11b motif
Sorted Z-values of sox11a+sox11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_22240052 | 6.16 |
ENSDART00000111109
|
crygm2d9
|
crystallin, gamma M2d9 |
| chr5_-_26118855 | 5.44 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr3_-_61205711 | 4.22 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr19_-_21832441 | 3.23 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr18_+_25653599 | 3.05 |
ENSDART00000007856
|
fkbp16
|
FK506 binding protein 16 |
| chr1_-_20928772 | 3.01 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr21_+_11401247 | 2.86 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr13_+_24755049 | 2.81 |
ENSDART00000134102
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr20_+_9128829 | 2.63 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr2_+_52232630 | 2.57 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr22_+_7439186 | 2.50 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr19_+_37509638 | 2.44 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr16_-_23346095 | 2.30 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr1_-_54765262 | 2.29 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr20_+_9128256 | 2.20 |
ENSDART00000163883
ENSDART00000183072 ENSDART00000187276 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr16_-_24598042 | 2.20 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr12_-_21834611 | 2.19 |
ENSDART00000179553
|
thrab
|
thyroid hormone receptor alpha b |
| chr11_+_30513656 | 2.18 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr2_-_32643738 | 2.17 |
ENSDART00000112452
|
si:dkeyp-73d8.9
|
si:dkeyp-73d8.9 |
| chr8_+_24861264 | 2.15 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr2_+_24177006 | 2.15 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr1_-_14234076 | 2.11 |
ENSDART00000040049
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr5_-_32308526 | 2.10 |
ENSDART00000193758
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr20_+_41664350 | 2.03 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr10_+_2587234 | 2.00 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr17_+_33340675 | 1.97 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr16_-_52540056 | 1.96 |
ENSDART00000188304
|
CR293507.1
|
|
| chr2_+_24177190 | 1.90 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr5_-_38342992 | 1.82 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr13_+_13693722 | 1.79 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr17_-_39185336 | 1.78 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr25_+_35056619 | 1.78 |
ENSDART00000154389
|
HIST1H4J
|
histone cluster 1 H4 family member j |
| chr23_+_40460333 | 1.75 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr8_-_14050758 | 1.70 |
ENSDART00000133922
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr21_-_41305748 | 1.68 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr11_+_39672874 | 1.64 |
ENSDART00000046663
ENSDART00000157659 |
camta1b
|
calmodulin binding transcription activator 1b |
| chr18_+_3579829 | 1.60 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr13_-_11644806 | 1.60 |
ENSDART00000169953
|
dctn1b
|
dynactin 1b |
| chr1_-_22512063 | 1.57 |
ENSDART00000031546
ENSDART00000190987 |
chrna6
|
cholinergic receptor, nicotinic, alpha 6 |
| chr23_-_17657348 | 1.54 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr8_-_10045043 | 1.53 |
ENSDART00000081917
|
si:dkey-8e10.3
|
si:dkey-8e10.3 |
| chr10_+_41668483 | 1.49 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr3_+_19299309 | 1.47 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr25_+_14087045 | 1.46 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr6_-_57539141 | 1.46 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr8_-_5220125 | 1.45 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr14_-_2187202 | 1.36 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr20_+_50852356 | 1.34 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr2_+_28672152 | 1.33 |
ENSDART00000157410
ENSDART00000169614 |
nadsyn1
|
NAD synthetase 1 |
| chr20_-_26039841 | 1.31 |
ENSDART00000179929
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr23_+_11669109 | 1.29 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr14_+_4151379 | 1.29 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr5_+_26795465 | 1.26 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr3_+_22270463 | 1.25 |
ENSDART00000104004
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr9_-_18568927 | 1.24 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr20_-_47731768 | 1.22 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr16_-_22006996 | 1.21 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr17_-_47090440 | 1.18 |
ENSDART00000163542
|
CABZ01056321.1
|
|
| chr13_-_40499296 | 1.15 |
ENSDART00000158338
|
CNNM1
|
Danio rerio cyclin and CBS domain divalent metal cation transport mediator 1 (cnnm1), mRNA. |
| chr2_-_37889111 | 1.15 |
ENSDART00000168939
ENSDART00000098529 |
mbl2
|
mannose binding lectin 2 |
| chr20_+_50115335 | 1.13 |
ENSDART00000031139
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr9_+_19039608 | 1.13 |
ENSDART00000055866
|
chmp2ba
|
charged multivesicular body protein 2Ba |
| chr6_+_54538948 | 1.12 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr20_-_35706379 | 1.08 |
ENSDART00000152847
|
si:dkey-30j22.1
|
si:dkey-30j22.1 |
| chr5_-_35301800 | 1.06 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr17_-_48915427 | 1.04 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr9_+_17984358 | 1.02 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr11_+_30282141 | 1.01 |
ENSDART00000122756
|
si:dkey-163f14.6
|
si:dkey-163f14.6 |
| chr21_+_26612777 | 0.98 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr6_+_39812475 | 0.96 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr16_-_33001153 | 0.95 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr2_-_1569250 | 0.94 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr2_-_26509057 | 0.92 |
ENSDART00000139169
ENSDART00000181777 |
best4
|
bestrophin 4 |
| chr9_+_38883388 | 0.88 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr20_+_54383838 | 0.88 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr15_-_2841677 | 0.86 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr24_+_22056386 | 0.86 |
ENSDART00000132390
|
ankrd33ba
|
ankyrin repeat domain 33ba |
| chr21_-_21530868 | 0.86 |
ENSDART00000174231
|
or133-9
|
odorant receptor, family H, subfamily 133, member 9 |
| chr22_+_7462997 | 0.85 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr20_+_37295006 | 0.84 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr5_+_37069605 | 0.83 |
ENSDART00000161977
|
si:dkeyp-110c7.4
|
si:dkeyp-110c7.4 |
| chr7_+_54642005 | 0.83 |
ENSDART00000171864
|
fgf19
|
fibroblast growth factor 19 |
| chr20_-_8304271 | 0.83 |
ENSDART00000179057
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr18_-_25177230 | 0.83 |
ENSDART00000013363
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr6_-_1768724 | 0.82 |
ENSDART00000162488
ENSDART00000163613 |
zgc:158417
|
zgc:158417 |
| chr10_+_38806719 | 0.81 |
ENSDART00000184416
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr6_-_59271436 | 0.80 |
ENSDART00000174588
|
ndufa4l2b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2b |
| chr12_+_32323098 | 0.80 |
ENSDART00000188722
|
ANKFN1
|
si:ch211-277e21.2 |
| chr24_-_41220538 | 0.80 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr23_+_4226341 | 0.80 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr15_-_40267485 | 0.78 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr19_+_2279051 | 0.78 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr19_+_32401278 | 0.77 |
ENSDART00000184353
|
atxn1a
|
ataxin 1a |
| chr18_-_3527686 | 0.75 |
ENSDART00000169049
|
capn5a
|
calpain 5a |
| chr11_-_25734417 | 0.75 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr17_-_43558494 | 0.74 |
ENSDART00000103830
|
nt5c1ab
|
5'-nucleotidase, cytosolic IAb |
| chr11_-_25733910 | 0.73 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr14_+_29200772 | 0.72 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr18_-_43884044 | 0.71 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr15_-_15357178 | 0.71 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr11_+_41243094 | 0.70 |
ENSDART00000191782
ENSDART00000169817 ENSDART00000162157 |
pax7a
|
paired box 7a |
| chr12_-_44010532 | 0.69 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr6_-_59271270 | 0.69 |
ENSDART00000126870
|
ndufa4l2b
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2b |
| chr17_+_47090497 | 0.68 |
ENSDART00000169038
ENSDART00000159292 |
zgc:103755
|
zgc:103755 |
| chr10_+_21650828 | 0.67 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr11_-_1409236 | 0.67 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr20_-_31067306 | 0.66 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr3_+_12861158 | 0.64 |
ENSDART00000171237
|
kcnj2b
|
potassium inwardly-rectifying channel, subfamily J, member 2b |
| chr1_-_36152131 | 0.64 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr1_+_10003193 | 0.64 |
ENSDART00000162675
|
trim2b
|
tripartite motif containing 2b |
| chr22_+_3156386 | 0.63 |
ENSDART00000161212
|
rpl36
|
ribosomal protein L36 |
| chr4_-_42391176 | 0.63 |
ENSDART00000189739
|
si:ch211-59d8.2
|
si:ch211-59d8.2 |
| chr7_+_19762595 | 0.62 |
ENSDART00000130347
|
si:dkey-9k7.3
|
si:dkey-9k7.3 |
| chr25_+_35058088 | 0.61 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr24_+_30521619 | 0.61 |
ENSDART00000162903
|
dpyda.3
|
dihydropyrimidine dehydrogenase a, tandem duplicate 3 |
| chr9_+_24008879 | 0.61 |
ENSDART00000190419
ENSDART00000191843 ENSDART00000148226 |
mlphb
|
melanophilin b |
| chr13_-_1349922 | 0.60 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr22_-_35961365 | 0.60 |
ENSDART00000188434
|
LO016987.2
|
|
| chr10_-_7857494 | 0.60 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr8_-_1838315 | 0.58 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr5_+_1983495 | 0.56 |
ENSDART00000179860
|
CT573103.2
|
|
| chr2_+_9061885 | 0.54 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr3_+_26267725 | 0.54 |
ENSDART00000131288
|
adap2
|
ArfGAP with dual PH domains 2 |
| chr11_-_891674 | 0.53 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr1_-_23596391 | 0.53 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr11_+_23760470 | 0.52 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr1_+_16397063 | 0.52 |
ENSDART00000159794
|
micu3a
|
mitochondrial calcium uptake family, member 3a |
| chr22_+_37902976 | 0.49 |
ENSDART00000169327
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr7_-_51727760 | 0.48 |
ENSDART00000174180
|
hdac8
|
histone deacetylase 8 |
| chr3_-_8399774 | 0.47 |
ENSDART00000190238
ENSDART00000113140 ENSDART00000132949 |
rbfox3b
|
RNA binding fox-1 homolog 3b |
| chr18_-_16392229 | 0.46 |
ENSDART00000189031
|
mgat4c
|
mgat4 family, member C |
| chr24_-_26302375 | 0.46 |
ENSDART00000130696
|
cops9
|
COP9 signalosome subunit 9 |
| chr15_-_38177768 | 0.46 |
ENSDART00000152404
|
si:dkey-24p1.7
|
si:dkey-24p1.7 |
| chr2_+_904362 | 0.46 |
ENSDART00000024967
|
ripk1l
|
receptor (TNFRSF)-interacting serine-threonine kinase 1, like |
| chr21_-_41025340 | 0.45 |
ENSDART00000148231
|
plac8l1
|
PLAC8-like 1 |
| chr19_-_31402429 | 0.45 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr15_-_38186537 | 0.41 |
ENSDART00000012872
|
chrna10a
|
cholinergic receptor, nicotinic, alpha 10a |
| chr16_+_36768674 | 0.41 |
ENSDART00000169208
ENSDART00000180470 |
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr19_-_47456787 | 0.40 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr5_-_57528943 | 0.40 |
ENSDART00000130320
|
pisd
|
phosphatidylserine decarboxylase |
| chr25_-_29087925 | 0.40 |
ENSDART00000171758
|
rpp25a
|
ribonuclease P and MRP subunit p25, a |
| chr10_+_6884123 | 0.40 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr2_-_2451181 | 0.40 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr11_-_16093018 | 0.39 |
ENSDART00000139309
ENSDART00000139819 |
SPATA1
|
si:dkey-205k8.5 |
| chr5_-_66301142 | 0.39 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr2_-_37532772 | 0.38 |
ENSDART00000133951
|
arhgef18a
|
rho/rac guanine nucleotide exchange factor (GEF) 18a |
| chr16_+_6957582 | 0.38 |
ENSDART00000111308
ENSDART00000187541 ENSDART00000149987 |
bbs9
|
Bardet-Biedl syndrome 9 |
| chr5_-_3118346 | 0.38 |
ENSDART00000167554
|
hsf5
|
heat shock transcription factor family member 5 |
| chr1_-_21297748 | 0.36 |
ENSDART00000142109
|
npy1r
|
neuropeptide Y receptor Y1 |
| chr5_-_13251907 | 0.35 |
ENSDART00000176774
ENSDART00000030553 |
top3b
|
DNA topoisomerase III beta |
| chr1_-_19079957 | 0.35 |
ENSDART00000141795
|
phox2ba
|
paired-like homeobox 2ba |
| chr21_-_30254185 | 0.35 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr16_-_44878245 | 0.35 |
ENSDART00000154391
ENSDART00000154925 ENSDART00000154697 |
arhgap33
|
Rho GTPase activating protein 33 |
| chr16_+_54263921 | 0.34 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr7_-_58729894 | 0.34 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr14_+_45970033 | 0.34 |
ENSDART00000047716
|
fermt3b
|
fermitin family member 3b |
| chr23_+_38953545 | 0.34 |
ENSDART00000184621
|
ATP9A
|
ATPase phospholipid transporting 9A (putative) |
| chr23_-_22650992 | 0.33 |
ENSDART00000079007
|
ca6
|
carbonic anhydrase VI |
| chr1_+_46948010 | 0.33 |
ENSDART00000132843
|
si:dkey-22n8.2
|
si:dkey-22n8.2 |
| chrM_+_6425 | 0.31 |
ENSDART00000093606
|
mt-co1
|
cytochrome c oxidase I, mitochondrial |
| chr19_+_26718074 | 0.31 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr6_-_31336317 | 0.31 |
ENSDART00000193927
|
dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr6_+_39098397 | 0.30 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr16_+_18535618 | 0.30 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chrM_+_12897 | 0.30 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr23_+_23022790 | 0.30 |
ENSDART00000189843
|
samd11
|
sterile alpha motif domain containing 11 |
| chr6_+_42820228 | 0.29 |
ENSDART00000185413
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr12_-_18408566 | 0.29 |
ENSDART00000078767
ENSDART00000152748 |
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr10_-_41907213 | 0.27 |
ENSDART00000167004
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr22_+_3238474 | 0.27 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr7_-_40145097 | 0.27 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr16_+_40217043 | 0.27 |
ENSDART00000191128
|
BX470182.1
|
|
| chr2_+_26240631 | 0.26 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr22_-_6361178 | 0.26 |
ENSDART00000150147
|
zgc:113298
|
zgc:113298 |
| chr15_-_20949692 | 0.25 |
ENSDART00000185548
|
tbcela
|
tubulin folding cofactor E-like a |
| chr5_-_41841892 | 0.25 |
ENSDART00000167089
|
si:dkey-65b12.6
|
si:dkey-65b12.6 |
| chr20_+_19793620 | 0.25 |
ENSDART00000136317
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr20_-_35012093 | 0.24 |
ENSDART00000062761
|
cnstb
|
consortin, connexin sorting protein b |
| chr18_+_5547185 | 0.24 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr10_-_39321367 | 0.23 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr15_-_45510977 | 0.22 |
ENSDART00000090596
|
fgf12b
|
fibroblast growth factor 12b |
| chr4_-_18635005 | 0.22 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr12_+_46696867 | 0.21 |
ENSDART00000152928
ENSDART00000153445 ENSDART00000123357 ENSDART00000152880 ENSDART00000123834 ENSDART00000174765 ENSDART00000189923 |
exoc7
|
exocyst complex component 7 |
| chr21_-_40317035 | 0.21 |
ENSDART00000143648
ENSDART00000013359 |
or101-1
|
odorant receptor, family B, subfamily 101, member 1 |
| chr16_-_17056630 | 0.19 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr12_-_20665164 | 0.19 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr19_-_1920613 | 0.19 |
ENSDART00000186176
|
si:ch211-149a19.3
|
si:ch211-149a19.3 |
| chr8_-_36140405 | 0.18 |
ENSDART00000182806
|
CT583723.2
|
|
| chr11_+_36355348 | 0.17 |
ENSDART00000145427
|
sypl2a
|
synaptophysin-like 2a |
| chr15_+_41932853 | 0.17 |
ENSDART00000183581
ENSDART00000157807 |
si:ch211-191a16.2
|
si:ch211-191a16.2 |
| chr23_-_6865946 | 0.16 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr8_-_38105053 | 0.14 |
ENSDART00000131546
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr13_-_45201300 | 0.14 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr6_+_4229360 | 0.14 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr16_-_15988320 | 0.13 |
ENSDART00000160883
|
CABZ01060453.1
|
|
| chr15_-_14467394 | 0.13 |
ENSDART00000191944
|
numbl
|
numb homolog (Drosophila)-like |
| chr8_+_8671229 | 0.12 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr9_-_5318873 | 0.12 |
ENSDART00000129308
|
ACVR1C
|
activin A receptor type 1C |
| chr20_+_16703050 | 0.11 |
ENSDART00000005244
|
kcnk13b
|
potassium channel, subfamily K, member 13b |
| chr12_-_37734973 | 0.10 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox11a+sox11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.7 | 2.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.7 | 2.0 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.4 | 1.3 | GO:0018315 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.4 | 1.3 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.3 | 1.3 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.3 | 2.9 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 1.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 5.4 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.2 | 1.8 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.2 | 2.2 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.2 | 0.8 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.2 | 1.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.2 | 0.5 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.2 | 0.6 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.1 | 1.5 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.1 | 0.7 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.1 | 0.6 | GO:0019860 | uracil catabolic process(GO:0006212) beta-alanine metabolic process(GO:0019482) beta-alanine biosynthetic process(GO:0019483) uracil metabolic process(GO:0019860) 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.1 | 0.7 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.1 | 0.3 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.4 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 1.3 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.1 | 0.9 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 1.1 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 2.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 1.3 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 2.1 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.1 | 3.0 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 1.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 1.8 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 1.6 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 0.3 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.7 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 7.0 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.1 | 1.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 0.8 | GO:1901571 | icosanoid transport(GO:0071715) fatty acid derivative transport(GO:1901571) |
| 0.0 | 0.5 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 1.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.2 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 1.5 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.0 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 2.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.3 | GO:0051967 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 1.0 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.1 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.1 | GO:0002676 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.5 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.0 | 0.8 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.8 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 2.8 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.2 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 2.1 | GO:0042391 | regulation of membrane potential(GO:0042391) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.5 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.5 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.4 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 0.8 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 1.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.6 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 3.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.9 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 4.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 6.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 12.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.4 | 1.3 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.4 | 3.0 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.4 | 1.5 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 5.6 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.2 | 0.9 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.2 | 2.0 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.2 | 3.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 1.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.6 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.1 | 1.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 1.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 1.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 1.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 2.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 2.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0070186 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.1 | 0.6 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.1 | 6.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 2.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 2.1 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 3.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 1.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.5 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.8 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 2.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 2.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 6.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 2.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 1.1 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 0.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.4 | 2.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 1.6 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 3.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.8 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |