Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
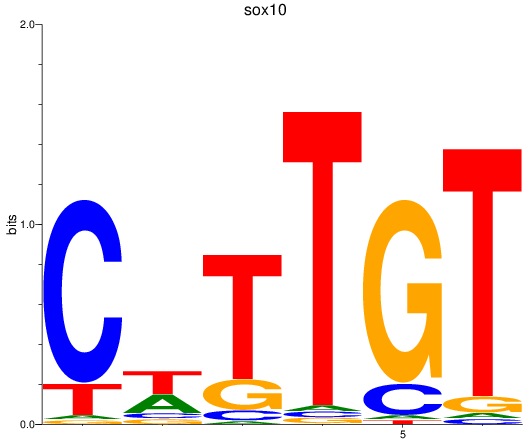
Results for sox10
Z-value: 0.92
Transcription factors associated with sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox10
|
ENSDARG00000077467 | SRY-box transcription factor 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox10 | dr11_v1_chr3_+_1492174_1492174 | 0.37 | 1.2e-01 | Click! |
Activity profile of sox10 motif
Sorted Z-values of sox10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_14098028 | 3.55 |
ENSDART00000018432
|
he1.2
|
hatching enzyme 1, tandem dupliate 2 |
| chr13_+_33651416 | 3.51 |
ENSDART00000180221
|
BX005372.1
|
|
| chr1_-_33645967 | 1.56 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr14_+_16036139 | 1.54 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr23_+_36144487 | 1.51 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr5_-_61652254 | 1.44 |
ENSDART00000097364
|
drl
|
draculin |
| chr6_+_45932276 | 1.38 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr23_-_21471022 | 1.35 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr2_-_42960353 | 1.26 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr22_-_10541372 | 1.25 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr11_+_3501669 | 1.21 |
ENSDART00000160808
|
pusl1
|
pseudouridylate synthase-like 1 |
| chr9_+_15893093 | 1.19 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr3_-_41535647 | 1.18 |
ENSDART00000153723
ENSDART00000154198 |
si:ch211-222n22.1
|
si:ch211-222n22.1 |
| chr19_-_47570672 | 1.18 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_-_18152407 | 1.18 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr3_-_16250527 | 1.16 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr14_+_25505468 | 1.13 |
ENSDART00000079016
|
thoc3
|
THO complex 3 |
| chr16_-_45917322 | 1.12 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr8_+_39802506 | 1.11 |
ENSDART00000018862
|
hnf1a
|
HNF1 homeobox a |
| chr24_+_22485710 | 1.11 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr7_+_22688781 | 1.09 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr15_-_36365840 | 1.05 |
ENSDART00000192926
|
si:dkey-23k10.3
|
si:dkey-23k10.3 |
| chr21_-_20342096 | 1.03 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr20_-_29498178 | 1.02 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr5_-_20205075 | 1.02 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr6_-_43092175 | 1.00 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr13_+_21677767 | 0.99 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr15_+_35691783 | 0.96 |
ENSDART00000183994
|
CT573366.1
|
|
| chr4_-_38877924 | 0.94 |
ENSDART00000177741
|
znf1127
|
zinc finger protein 1127 |
| chr19_-_17208728 | 0.93 |
ENSDART00000151228
|
stmn1a
|
stathmin 1a |
| chr15_-_3736149 | 0.93 |
ENSDART00000182986
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr13_+_18321140 | 0.92 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr22_-_15587360 | 0.91 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr21_-_25741411 | 0.91 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr6_-_54290227 | 0.90 |
ENSDART00000050483
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr22_-_18546241 | 0.89 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr24_+_18714212 | 0.89 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr5_+_67662430 | 0.89 |
ENSDART00000137700
ENSDART00000142586 |
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr3_-_26184018 | 0.88 |
ENSDART00000191604
|
si:ch211-11k18.4
|
si:ch211-11k18.4 |
| chr16_+_26732086 | 0.87 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr6_-_6993046 | 0.87 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr4_+_63088692 | 0.87 |
ENSDART00000168872
|
zgc:173714
|
zgc:173714 |
| chr7_+_24814866 | 0.86 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr22_-_10541712 | 0.86 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_-_31258459 | 0.85 |
ENSDART00000177928
|
LO017965.1
|
|
| chr14_+_989733 | 0.85 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr17_-_16422654 | 0.85 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr7_-_38689562 | 0.85 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr17_+_15535501 | 0.84 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr7_+_22982201 | 0.84 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr18_-_37241080 | 0.83 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr4_+_25607743 | 0.83 |
ENSDART00000028297
|
acot14
|
acyl-CoA thioesterase 14 |
| chr18_-_35407695 | 0.82 |
ENSDART00000191845
ENSDART00000141703 |
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr19_+_43017931 | 0.81 |
ENSDART00000132213
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr16_-_32233463 | 0.81 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr23_-_30727596 | 0.81 |
ENSDART00000060193
|
thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr18_-_25051846 | 0.79 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr25_+_14400704 | 0.78 |
ENSDART00000067235
|
ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr3_-_31069776 | 0.77 |
ENSDART00000167462
|
elob
|
elongin B |
| chr10_+_44984946 | 0.77 |
ENSDART00000182520
|
h2afvb
|
H2A histone family, member Vb |
| chr13_+_46941930 | 0.77 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr24_+_37799818 | 0.77 |
ENSDART00000131975
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
| chr4_+_5334439 | 0.77 |
ENSDART00000180644
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr2_-_25140022 | 0.77 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr15_-_31514818 | 0.77 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr8_-_16697912 | 0.77 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr4_-_20108833 | 0.76 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr11_-_41220794 | 0.75 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr15_-_47822597 | 0.75 |
ENSDART00000193236
ENSDART00000161391 |
CZQB01095947.1
|
|
| chr13_+_33298338 | 0.75 |
ENSDART00000131892
ENSDART00000143895 |
iqcc
|
IQ motif containing C |
| chr21_-_20341836 | 0.75 |
ENSDART00000176689
|
rbp4l
|
retinol binding protein 4, like |
| chr19_-_27588842 | 0.74 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr19_+_3215466 | 0.74 |
ENSDART00000181288
|
zgc:86598
|
zgc:86598 |
| chr14_-_38827442 | 0.73 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr4_-_71110826 | 0.73 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr9_-_18911608 | 0.73 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr21_-_5077715 | 0.72 |
ENSDART00000081954
|
haus1
|
HAUS augmin-like complex, subunit 1 |
| chr3_-_29977495 | 0.71 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr1_-_53277660 | 0.71 |
ENSDART00000014232
|
znf330
|
zinc finger protein 330 |
| chr18_+_31117136 | 0.70 |
ENSDART00000138403
|
tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr7_+_22981909 | 0.69 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr2_+_35595454 | 0.69 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr9_+_8396755 | 0.69 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr4_+_69577362 | 0.68 |
ENSDART00000160913
|
znf1030
|
zinc finger protein 1030 |
| chr21_-_41065369 | 0.68 |
ENSDART00000143749
|
larsb
|
leucyl-tRNA synthetase b |
| chr14_-_4170654 | 0.68 |
ENSDART00000188347
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr2_+_10821579 | 0.68 |
ENSDART00000179694
|
glmna
|
glomulin, FKBP associated protein a |
| chr8_+_23356264 | 0.67 |
ENSDART00000145062
|
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr23_+_36063599 | 0.67 |
ENSDART00000103147
|
hoxc12a
|
homeobox C12a |
| chr11_+_6882049 | 0.66 |
ENSDART00000075998
|
klhl26
|
kelch-like family member 26 |
| chr20_+_31217495 | 0.65 |
ENSDART00000020252
|
pdia6
|
protein disulfide isomerase family A, member 6 |
| chr13_+_51579851 | 0.65 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr1_+_35862550 | 0.65 |
ENSDART00000132118
|
si:ch211-194g2.4
|
si:ch211-194g2.4 |
| chr5_+_25681174 | 0.65 |
ENSDART00000134773
|
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr16_+_15114645 | 0.65 |
ENSDART00000158483
|
mtbp
|
MDM2 binding protein |
| chr9_+_23003208 | 0.65 |
ENSDART00000021060
|
eaf2
|
ELL associated factor 2 |
| chr21_-_26495700 | 0.65 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr21_+_44684577 | 0.64 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr8_+_39619087 | 0.63 |
ENSDART00000134822
|
msi1
|
musashi RNA-binding protein 1 |
| chr5_+_32141790 | 0.63 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr24_+_11083146 | 0.63 |
ENSDART00000009473
|
zfand1
|
zinc finger, AN1-type domain 1 |
| chr17_+_24064014 | 0.62 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr20_+_18703454 | 0.62 |
ENSDART00000152342
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr20_+_25586099 | 0.61 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr2_-_21170517 | 0.60 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr16_+_35401543 | 0.60 |
ENSDART00000171608
|
rab42b
|
RAB42, member RAS oncogene family |
| chr19_+_1872794 | 0.60 |
ENSDART00000013217
|
snrpd1
|
small nuclear ribonucleoprotein D1 polypeptide |
| chr5_-_41645058 | 0.59 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr11_+_37720756 | 0.59 |
ENSDART00000173337
|
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr5_-_52215926 | 0.59 |
ENSDART00000163973
ENSDART00000193602 |
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr17_+_24851951 | 0.58 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr20_+_11734029 | 0.58 |
ENSDART00000179959
|
si:ch211-155o21.3
|
si:ch211-155o21.3 |
| chr13_+_43247936 | 0.57 |
ENSDART00000126850
ENSDART00000165331 |
smoc2
|
SPARC related modular calcium binding 2 |
| chr5_+_26121393 | 0.57 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr16_+_9762261 | 0.57 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr2_-_56649883 | 0.57 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr4_+_69375564 | 0.57 |
ENSDART00000150334
|
si:dkey-246j6.1
|
si:dkey-246j6.1 |
| chr3_-_26204867 | 0.57 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr23_+_36340520 | 0.57 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr6_+_36459897 | 0.57 |
ENSDART00000104248
ENSDART00000145753 |
lrrc15
|
leucine rich repeat containing 15 |
| chr14_-_38828057 | 0.57 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr13_-_35459928 | 0.56 |
ENSDART00000144109
|
slx4ip
|
SLX4 interacting protein |
| chr14_+_36220479 | 0.56 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr21_-_2310064 | 0.56 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr2_-_30912922 | 0.56 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr19_+_18903533 | 0.56 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_42213285 | 0.55 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr15_+_834697 | 0.55 |
ENSDART00000154861
ENSDART00000160694 |
si:dkey-7i4.14
zgc:174573
|
si:dkey-7i4.14 zgc:174573 |
| chr14_-_24101897 | 0.55 |
ENSDART00000143695
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr19_+_13410903 | 0.54 |
ENSDART00000165033
ENSDART00000168672 |
srsf4
|
serine/arginine-rich splicing factor 4 |
| chr22_-_17489040 | 0.54 |
ENSDART00000141286
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr5_+_9134187 | 0.54 |
ENSDART00000132240
|
pimr123
|
Pim proto-oncogene, serine/threonine kinase, related 123 |
| chr17_+_43595692 | 0.54 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr8_+_21133307 | 0.54 |
ENSDART00000056405
|
magoh
|
mago homolog, exon junction complex core component |
| chr3_+_41922114 | 0.54 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr23_-_10175898 | 0.53 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr16_-_25699554 | 0.53 |
ENSDART00000155899
ENSDART00000114495 |
ddx61
|
DEAD (Asp-Glu-Ala-Asp) box helicase 61 |
| chr23_-_3758637 | 0.53 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr11_-_7147540 | 0.53 |
ENSDART00000143942
|
bmp7a
|
bone morphogenetic protein 7a |
| chr17_-_6599484 | 0.53 |
ENSDART00000156927
|
ANKRD66
|
si:ch211-189e2.2 |
| chr16_-_27566552 | 0.52 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr5_+_25680845 | 0.52 |
ENSDART00000139701
ENSDART00000009952 |
zfand5a
|
zinc finger, AN1-type domain 5a |
| chr16_+_14201401 | 0.52 |
ENSDART00000113679
|
dap3
|
death associated protein 3 |
| chr21_+_7131970 | 0.51 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr6_-_7020162 | 0.51 |
ENSDART00000148982
|
bin1b
|
bridging integrator 1b |
| chr17_-_49438873 | 0.51 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr13_+_22675802 | 0.51 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr17_-_51222510 | 0.51 |
ENSDART00000166687
|
psen1
|
presenilin 1 |
| chr14_+_18782610 | 0.51 |
ENSDART00000164468
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr5_+_53482597 | 0.51 |
ENSDART00000180333
|
BX323994.1
|
|
| chr2_-_38080075 | 0.51 |
ENSDART00000056544
|
tox4a
|
TOX high mobility group box family member 4 a |
| chr7_+_19835569 | 0.50 |
ENSDART00000149812
|
ovol1a
|
ovo-like zinc finger 1a |
| chr25_-_27541621 | 0.49 |
ENSDART00000130678
|
spam1
|
sperm adhesion molecule 1 |
| chr21_-_25756119 | 0.49 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr7_+_10562118 | 0.49 |
ENSDART00000185188
ENSDART00000168801 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr23_-_36446307 | 0.49 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr12_-_30777540 | 0.49 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr19_+_26714949 | 0.49 |
ENSDART00000188540
|
zgc:100906
|
zgc:100906 |
| chr21_-_40348790 | 0.49 |
ENSDART00000178123
|
CR847523.1
|
|
| chr5_-_56948058 | 0.48 |
ENSDART00000083074
ENSDART00000191028 |
si:ch211-127d4.3
|
si:ch211-127d4.3 |
| chr2_+_30513887 | 0.48 |
ENSDART00000137048
|
march6
|
membrane-associated ring finger (C3HC4) 6 |
| chr19_-_3876877 | 0.48 |
ENSDART00000163711
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr14_-_10387377 | 0.48 |
ENSDART00000145118
|
commd5
|
COMM domain containing 5 |
| chr18_-_2668698 | 0.48 |
ENSDART00000157510
|
relt
|
RELT, TNF receptor |
| chr16_+_35535375 | 0.47 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr16_+_19637384 | 0.47 |
ENSDART00000184773
ENSDART00000191895 ENSDART00000182020 ENSDART00000135359 |
macc1
|
metastasis associated in colon cancer 1 |
| chr11_+_13528437 | 0.47 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr1_-_52790724 | 0.47 |
ENSDART00000139577
ENSDART00000100937 |
patl1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr17_+_24848976 | 0.47 |
ENSDART00000062917
|
cx35.4
|
connexin 35.4 |
| chr21_+_22878991 | 0.47 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr25_+_15994100 | 0.47 |
ENSDART00000144723
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr2_-_30912307 | 0.46 |
ENSDART00000188653
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr18_-_40773413 | 0.46 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr5_-_28625515 | 0.46 |
ENSDART00000190782
ENSDART00000179736 ENSDART00000131729 |
tnc
|
tenascin C |
| chr7_+_73819078 | 0.46 |
ENSDART00000169756
|
FP236812.1
|
Histone H2B 1/2 |
| chr15_-_18218227 | 0.46 |
ENSDART00000156332
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr9_-_7683799 | 0.45 |
ENSDART00000102713
|
si:ch73-199e17.1
|
si:ch73-199e17.1 |
| chr4_+_62184754 | 0.45 |
ENSDART00000168844
|
si:dkeyp-35e5.9
|
si:dkeyp-35e5.9 |
| chr21_-_28920245 | 0.45 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr25_+_16880990 | 0.45 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr21_+_19347655 | 0.45 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr13_-_1151410 | 0.45 |
ENSDART00000007231
|
psmb1
|
proteasome subunit beta 1 |
| chr13_+_15933168 | 0.45 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr23_+_21492151 | 0.45 |
ENSDART00000025487
|
icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr9_+_34151367 | 0.44 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr5_+_63329608 | 0.44 |
ENSDART00000139180
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr5_+_3892551 | 0.44 |
ENSDART00000134396
|
rpain
|
RPA interacting protein |
| chr20_+_34129465 | 0.44 |
ENSDART00000168804
ENSDART00000023211 |
pla2g4ab
|
phospholipase A2, group IVAb (cytosolic, calcium-dependent) |
| chr10_+_9550419 | 0.44 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr19_+_16015881 | 0.44 |
ENSDART00000187135
|
ctps1a
|
CTP synthase 1a |
| chr20_-_22476255 | 0.44 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr10_+_35422802 | 0.43 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr6_-_39313027 | 0.43 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr19_+_43017628 | 0.43 |
ENSDART00000182608
ENSDART00000015632 |
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr19_+_42432625 | 0.43 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr2_-_17115256 | 0.43 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr12_-_3077395 | 0.43 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr11_+_37612214 | 0.42 |
ENSDART00000172899
ENSDART00000077496 |
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr6_-_21091948 | 0.42 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr9_-_9242777 | 0.42 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.6 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.3 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 0.9 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.3 | 1.0 | GO:0055130 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 1.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 2.2 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.2 | 1.9 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 0.8 | GO:1904667 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.2 | 0.8 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.2 | 0.6 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.2 | 0.8 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.5 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.5 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.1 | 0.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 1.0 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.6 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.1 | 0.8 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 1.4 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.3 | GO:2000639 | response to sterol(GO:0036314) cellular response to sterol(GO:0036315) SREBP-SCAP complex retention in endoplasmic reticulum(GO:0036316) regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.6 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 1.1 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.1 | 0.5 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.4 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.3 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.1 | 1.4 | GO:0021594 | rhombomere formation(GO:0021594) |
| 0.1 | 0.4 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 1.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.1 | 1.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.7 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 1.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.2 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.1 | 1.1 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.1 | 0.3 | GO:0046102 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.4 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.4 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.9 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.3 | GO:0097638 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 1.0 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.4 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.6 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 1.2 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.8 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.0 | 0.4 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.8 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.0 | 0.6 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:2000406 | positive regulation of lymphocyte migration(GO:2000403) positive regulation of T cell migration(GO:2000406) |
| 0.0 | 0.4 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 1.1 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.2 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.3 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.6 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 1.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.1 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 2.1 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 0.6 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.4 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0046324 | regulation of glucose import(GO:0046324) |
| 0.0 | 0.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 | 0.7 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.2 | GO:0018120 | peptidyl-arginine ADP-ribosylation(GO:0018120) |
| 0.0 | 0.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.6 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.5 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 1.0 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.6 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.0 | GO:0006354 | DNA-templated transcription, elongation(GO:0006354) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.8 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 1.4 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.0 | 0.4 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.1 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.1 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.6 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.9 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.6 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.9 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.3 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.2 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.0 | 0.3 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 1.0 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.8 | GO:0001666 | response to hypoxia(GO:0001666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 0.8 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.2 | 1.1 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.8 | GO:0030891 | VCB complex(GO:0030891) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 0.4 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 0.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.6 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 0.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 0.6 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.2 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.1 | 0.7 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.1 | 2.3 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.4 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.6 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.6 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.4 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.9 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.6 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 1.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 1.9 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.0 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.2 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.0 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.9 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.3 | 0.8 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.3 | 1.0 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.3 | 1.8 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.2 | 1.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 1.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 0.6 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.8 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.2 | 0.7 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.2 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 0.8 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.1 | 0.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.9 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.4 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.1 | 0.6 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.1 | 0.4 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.3 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 0.4 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.4 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 1.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.2 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.1 | 0.3 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 0.9 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.6 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 1.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.3 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.1 | 0.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.5 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.4 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.1 | 0.7 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.8 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.4 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.4 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.3 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.2 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.0 | 0.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.5 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.6 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 1.5 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.9 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.4 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 1.6 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 2.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.6 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.8 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 2.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.6 | REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | Genes involved in Cleavage of Growing Transcript in the Termination Region |
| 0.1 | 0.4 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.4 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.6 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 0.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.1 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.8 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |