Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
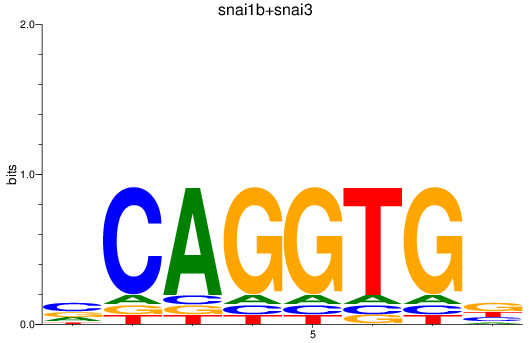
Results for snai1b+snai3
Z-value: 0.70
Transcription factors associated with snai1b+snai3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai3
|
ENSDARG00000031243 | snail family zinc finger 3 |
|
snai1b
|
ENSDARG00000046019 | snail family zinc finger 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai1b | dr11_v1_chr23_-_2513300_2513300 | 0.38 | 1.1e-01 | Click! |
| snai3 | dr11_v1_chr7_+_55112922_55112922 | -0.13 | 5.9e-01 | Click! |
Activity profile of snai1b+snai3 motif
Sorted Z-values of snai1b+snai3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_21465111 | 2.73 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr19_-_35439237 | 1.75 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr14_-_26482096 | 1.11 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr22_+_22438783 | 1.01 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr17_-_11151655 | 1.01 |
ENSDART00000156383
|
CU179699.1
|
|
| chr7_+_20917966 | 0.97 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr6_-_130849 | 0.96 |
ENSDART00000108710
|
LRRC8E
|
si:zfos-323e3.4 |
| chr7_-_50367642 | 0.94 |
ENSDART00000134941
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr13_+_35955562 | 0.94 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr3_+_14641962 | 0.92 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr8_+_247163 | 0.90 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr4_-_78026285 | 0.85 |
ENSDART00000168273
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr9_+_331425 | 0.84 |
ENSDART00000170312
|
actr5
|
ARP5 actin related protein 5 homolog |
| chr14_-_31087830 | 0.83 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr22_-_26251563 | 0.82 |
ENSDART00000060888
ENSDART00000142821 |
ccdc130
|
coiled-coil domain containing 130 |
| chr6_-_10788065 | 0.82 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr6_-_10728921 | 0.80 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr14_+_34514336 | 0.78 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr17_+_1360192 | 0.76 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr6_-_10728057 | 0.73 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr2_-_30055432 | 0.72 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr13_-_21672131 | 0.72 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr25_-_16076257 | 0.71 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr10_-_94184 | 0.69 |
ENSDART00000146125
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr14_-_38865800 | 0.68 |
ENSDART00000173047
|
gsr
|
glutathione reductase |
| chr9_-_28255029 | 0.65 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr13_-_39254 | 0.63 |
ENSDART00000093222
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr20_-_21672970 | 0.62 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr25_+_5249513 | 0.61 |
ENSDART00000126814
|
CABZ01039863.1
|
|
| chr19_+_142270 | 0.60 |
ENSDART00000160201
|
tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b |
| chr12_-_11294979 | 0.60 |
ENSDART00000148850
|
get4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr17_+_45639247 | 0.59 |
ENSDART00000153669
|
vcpkmt
|
valosin containing protein lysine (K) methyltransferase |
| chr5_-_22052852 | 0.57 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr5_+_53482597 | 0.55 |
ENSDART00000180333
|
BX323994.1
|
|
| chr2_-_722156 | 0.55 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr21_-_43666420 | 0.53 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr20_-_9436521 | 0.53 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr2_-_42628028 | 0.52 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr13_-_9119867 | 0.51 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr13_+_2894536 | 0.50 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr14_-_15155384 | 0.50 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr13_-_36579086 | 0.50 |
ENSDART00000146671
|
lgals3a
|
lectin, galactoside binding soluble 3a |
| chr5_+_6670945 | 0.49 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr23_-_29824146 | 0.48 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr13_+_23157053 | 0.46 |
ENSDART00000162359
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_-_43743700 | 0.46 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr2_+_26237322 | 0.45 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr22_+_2417105 | 0.44 |
ENSDART00000106415
|
zgc:113220
|
zgc:113220 |
| chr3_-_34052882 | 0.44 |
ENSDART00000151463
|
ighv11-1
|
immunoglobulin heavy variable 11-1 |
| chr15_+_1397811 | 0.43 |
ENSDART00000102125
|
schip1
|
schwannomin interacting protein 1 |
| chr10_+_1052591 | 0.41 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr1_+_524717 | 0.40 |
ENSDART00000102421
ENSDART00000184473 |
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr7_-_50367326 | 0.39 |
ENSDART00000141926
|
prc1b
|
protein regulator of cytokinesis 1b |
| chr12_+_48784731 | 0.38 |
ENSDART00000158348
|
zmiz1b
|
zinc finger, MIZ-type containing 1b |
| chr24_+_19578935 | 0.36 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr22_-_17256573 | 0.36 |
ENSDART00000136119
ENSDART00000062891 |
nphs2
|
nephrosis 2, idiopathic, steroid-resistant (podocin) |
| chr18_+_27821856 | 0.35 |
ENSDART00000131712
|
si:ch211-222m18.4
|
si:ch211-222m18.4 |
| chr23_+_32039386 | 0.33 |
ENSDART00000133801
|
mylk2
|
myosin light chain kinase 2 |
| chr12_-_2869565 | 0.33 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr20_-_23426339 | 0.32 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr17_+_24590177 | 0.32 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr22_-_8910567 | 0.32 |
ENSDART00000187896
ENSDART00000188511 ENSDART00000175764 |
FO393424.2
|
|
| chr21_-_41873584 | 0.31 |
ENSDART00000188089
|
endou2
|
endonuclease, polyU-specific 2 |
| chr24_+_9372292 | 0.31 |
ENSDART00000082422
ENSDART00000191127 ENSDART00000180510 |
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr18_-_17531483 | 0.30 |
ENSDART00000061000
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr1_-_43892349 | 0.28 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr22_-_17677947 | 0.27 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr22_+_20560041 | 0.26 |
ENSDART00000135490
|
nfil3-4
|
nuclear factor, interleukin 3 regulated, member 4 |
| chr3_+_24190207 | 0.26 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr25_+_5012791 | 0.25 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr25_+_27744293 | 0.23 |
ENSDART00000103519
ENSDART00000149456 |
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr10_-_13116337 | 0.23 |
ENSDART00000164568
|
musk
|
muscle, skeletal, receptor tyrosine kinase |
| chr24_+_40860320 | 0.22 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr17_+_50655619 | 0.22 |
ENSDART00000184917
ENSDART00000167638 |
ddhd1a
|
DDHD domain containing 1a |
| chr21_+_30794351 | 0.22 |
ENSDART00000139486
|
zgc:158225
|
zgc:158225 |
| chr19_-_3106447 | 0.22 |
ENSDART00000137987
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr22_-_209741 | 0.21 |
ENSDART00000171954
|
ora1
|
olfactory receptor class A related 1 |
| chr7_-_30926030 | 0.21 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr13_-_11800281 | 0.21 |
ENSDART00000079392
|
zmp:0000000662
|
zmp:0000000662 |
| chr18_+_33580660 | 0.20 |
ENSDART00000139117
|
si:dkey-47k20.4
|
si:dkey-47k20.4 |
| chr9_-_11549379 | 0.20 |
ENSDART00000187074
|
fev
|
FEV (ETS oncogene family) |
| chr10_-_33588106 | 0.19 |
ENSDART00000155634
|
eva1c
|
eva-1 homolog C (C. elegans) |
| chr12_+_16953415 | 0.18 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr14_-_28052474 | 0.18 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr8_+_20140321 | 0.18 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr15_+_784149 | 0.17 |
ENSDART00000155114
|
znf970
|
zinc finger protein 970 |
| chr7_+_30926289 | 0.16 |
ENSDART00000173518
|
si:dkey-1h4.4
|
si:dkey-1h4.4 |
| chr2_-_10896745 | 0.15 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr10_-_8358396 | 0.12 |
ENSDART00000059322
|
csgalnact1a
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1a |
| chr21_+_19858627 | 0.11 |
ENSDART00000147010
|
fybb
|
FYN binding protein b |
| chr18_+_45666489 | 0.11 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr3_+_23029934 | 0.11 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr7_-_58867188 | 0.11 |
ENSDART00000187006
|
CU681855.1
|
|
| chr7_+_58751504 | 0.10 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr21_-_18648861 | 0.10 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr9_+_24708653 | 0.09 |
ENSDART00000065136
|
CT030712.1
|
|
| chr24_-_25184553 | 0.09 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr10_-_38468847 | 0.08 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr22_-_3595439 | 0.08 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr7_+_42328578 | 0.07 |
ENSDART00000149082
|
phkb
|
phosphorylase kinase, beta |
| chr18_+_6641542 | 0.07 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr4_-_12790886 | 0.06 |
ENSDART00000182535
ENSDART00000067131 ENSDART00000186426 |
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr11_-_8782871 | 0.02 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr6_-_37749711 | 0.01 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr2_+_9536021 | 0.01 |
ENSDART00000187357
|
CU929379.1
|
|
| chr4_-_36476889 | 0.01 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr3_-_42016693 | 0.01 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr8_+_8196087 | 0.01 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr11_-_43473824 | 0.00 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr20_-_1314355 | 0.00 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai1b+snai3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.2 | 0.9 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.2 | 0.8 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.2 | 1.0 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 1.3 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.5 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 2.7 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.7 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 1.1 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.5 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.6 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.7 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.6 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.8 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.3 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.6 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0032640 | tumor necrosis factor production(GO:0032640) tumor necrosis factor superfamily cytokine production(GO:0071706) |
| 0.0 | 0.4 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 1.6 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.8 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 1.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.6 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.1 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.8 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.9 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 0.5 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.7 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.2 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.7 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.4 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 2.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.8 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |