Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
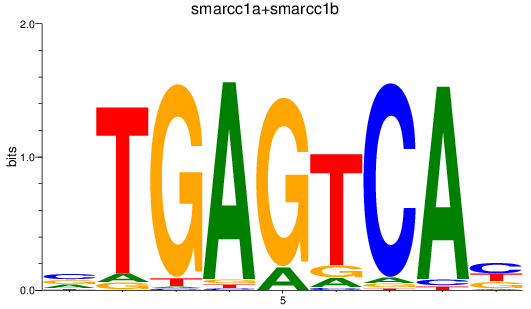
Results for smarcc1a+smarcc1b
Z-value: 1.01
Transcription factors associated with smarcc1a+smarcc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc1a
|
ENSDARG00000017397 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
|
smarcc1b
|
ENSDARG00000098919 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc1b | dr11_v1_chr19_-_19379084_19379084 | -0.27 | 2.6e-01 | Click! |
| smarcc1a | dr11_v1_chr16_-_42461263_42461263 | -0.22 | 3.6e-01 | Click! |
Activity profile of smarcc1a+smarcc1b motif
Sorted Z-values of smarcc1a+smarcc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_47115787 | 5.67 |
ENSDART00000192601
|
CABZ01079081.1
|
|
| chr3_-_4303262 | 4.16 |
ENSDART00000112819
|
si:dkey-73p2.2
|
si:dkey-73p2.2 |
| chr7_+_35068036 | 3.71 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr1_-_59252973 | 3.67 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr16_+_18974064 | 3.48 |
ENSDART00000079248
|
slc6a19b
|
solute carrier family 6 (neutral amino acid transporter), member 19b |
| chr24_-_40009446 | 3.16 |
ENSDART00000087422
|
aoc1
|
amine oxidase, copper containing 1 |
| chr11_+_24002503 | 2.62 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr19_-_21832441 | 2.58 |
ENSDART00000151272
ENSDART00000151442 ENSDART00000150168 ENSDART00000148797 ENSDART00000128196 ENSDART00000149259 ENSDART00000052556 ENSDART00000149658 ENSDART00000149639 ENSDART00000148424 |
mbpa
|
myelin basic protein a |
| chr2_-_293793 | 2.54 |
ENSDART00000082091
|
si:ch73-40a17.3
|
si:ch73-40a17.3 |
| chr12_-_30841679 | 2.51 |
ENSDART00000105594
|
crygmx
|
crystallin, gamma MX |
| chr6_-_13783604 | 2.37 |
ENSDART00000149536
ENSDART00000041269 ENSDART00000150102 |
cryba2a
|
crystallin, beta A2a |
| chr16_+_14033121 | 2.36 |
ENSDART00000135844
|
rusc1
|
RUN and SH3 domain containing 1 |
| chr8_+_43053519 | 2.35 |
ENSDART00000147178
|
prnpa
|
prion protein a |
| chr6_-_12588044 | 2.30 |
ENSDART00000047896
|
slc15a1b
|
solute carrier family 15 (oligopeptide transporter), member 1b |
| chr5_-_20678300 | 2.11 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr5_-_50992690 | 2.09 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr18_-_5692292 | 2.09 |
ENSDART00000121503
|
cplx3b
|
complexin 3b |
| chr15_+_28989573 | 1.94 |
ENSDART00000076648
|
clip3
|
CAP-GLY domain containing linker protein 3 |
| chr9_+_56422311 | 1.89 |
ENSDART00000171958
|
gpr39
|
G protein-coupled receptor 39 |
| chr2_-_3437862 | 1.77 |
ENSDART00000053012
|
cyp8b1
|
cytochrome P450, family 8, subfamily B, polypeptide 1 |
| chr11_-_43104475 | 1.76 |
ENSDART00000125368
|
acyp2
|
acylphosphatase 2, muscle type |
| chr24_+_35947077 | 1.75 |
ENSDART00000173406
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr9_-_12493628 | 1.71 |
ENSDART00000180832
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr5_-_22501663 | 1.68 |
ENSDART00000133174
|
si:dkey-27p18.5
|
si:dkey-27p18.5 |
| chr22_-_10110959 | 1.67 |
ENSDART00000031005
ENSDART00000147580 |
gls2b
|
glutaminase 2b (liver, mitochondrial) |
| chr2_-_155270 | 1.67 |
ENSDART00000131177
|
adcy1b
|
adenylate cyclase 1b |
| chr3_-_61185746 | 1.65 |
ENSDART00000028219
|
pvalb4
|
parvalbumin 4 |
| chr19_-_703898 | 1.65 |
ENSDART00000181096
ENSDART00000121462 |
slc6a19a.2
|
solute carrier family 6 (neutral amino acid transporter), member 19a, tandem duplicate 2 |
| chr4_+_74131530 | 1.65 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr23_-_40250886 | 1.61 |
ENSDART00000041912
|
tgm1l3
|
transglutaminase 1 like 3 |
| chr16_+_49005321 | 1.59 |
ENSDART00000160919
|
CABZ01068499.1
|
|
| chr22_+_1049367 | 1.58 |
ENSDART00000065380
|
camk1ga
|
calcium/calmodulin-dependent protein kinase IGa |
| chr1_+_9071324 | 1.58 |
ENSDART00000184194
|
BX004779.2
|
|
| chr1_+_45323142 | 1.58 |
ENSDART00000132210
|
emp1
|
epithelial membrane protein 1 |
| chr16_-_36834505 | 1.54 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr21_+_6328801 | 1.53 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr1_-_54765262 | 1.53 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr2_+_46589798 | 1.49 |
ENSDART00000128457
ENSDART00000145347 |
ephb1
|
EPH receptor B1 |
| chr17_+_22311413 | 1.49 |
ENSDART00000151929
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr12_+_22607761 | 1.49 |
ENSDART00000153112
|
si:dkey-219e21.2
|
si:dkey-219e21.2 |
| chr5_+_21931124 | 1.49 |
ENSDART00000137627
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr15_-_45674213 | 1.46 |
ENSDART00000080072
|
CABZ01067232.1
|
|
| chr7_+_34492744 | 1.46 |
ENSDART00000109635
ENSDART00000173844 |
calml4a
|
calmodulin-like 4a |
| chr23_-_270847 | 1.45 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr1_+_9004719 | 1.44 |
ENSDART00000006211
ENSDART00000137211 |
prkcba
|
protein kinase C, beta a |
| chr21_-_42100471 | 1.44 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr11_-_10770053 | 1.44 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr22_-_29191152 | 1.42 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr3_+_3810919 | 1.42 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr22_+_19247255 | 1.36 |
ENSDART00000144053
|
si:dkey-21e2.10
|
si:dkey-21e2.10 |
| chr5_+_62356304 | 1.35 |
ENSDART00000148381
|
aspa
|
aspartoacylase |
| chr16_-_13818061 | 1.33 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr13_-_293250 | 1.33 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr13_+_23988442 | 1.31 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr3_+_4346854 | 1.30 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr5_-_37935607 | 1.29 |
ENSDART00000141233
ENSDART00000084868 ENSDART00000125857 |
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr23_-_7799184 | 1.29 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr22_-_24297510 | 1.28 |
ENSDART00000163297
|
si:ch211-117l17.6
|
si:ch211-117l17.6 |
| chr22_-_6562618 | 1.27 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr9_+_33154841 | 1.27 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr12_+_47280839 | 1.25 |
ENSDART00000187574
|
RYR2
|
ryanodine receptor 2 |
| chr21_+_13353263 | 1.24 |
ENSDART00000114677
|
si:ch73-62l21.1
|
si:ch73-62l21.1 |
| chr4_-_789645 | 1.21 |
ENSDART00000164441
|
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr10_-_105100 | 1.21 |
ENSDART00000145716
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr24_-_11309477 | 1.20 |
ENSDART00000137257
|
myrip
|
myosin VIIA and Rab interacting protein |
| chr13_+_1430094 | 1.18 |
ENSDART00000169888
|
si:ch211-165e15.1
|
si:ch211-165e15.1 |
| chr19_+_1510971 | 1.18 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr22_+_28337204 | 1.15 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr1_-_45920632 | 1.15 |
ENSDART00000140890
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr9_-_34915351 | 1.14 |
ENSDART00000100728
ENSDART00000139608 |
upf3a
|
UPF3A, regulator of nonsense mediated mRNA decay |
| chr18_+_15841449 | 1.14 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr4_-_53047883 | 1.14 |
ENSDART00000157496
|
si:dkey-201g16.5
|
si:dkey-201g16.5 |
| chr1_+_6640437 | 1.13 |
ENSDART00000147638
|
si:ch211-93g23.2
|
si:ch211-93g23.2 |
| chr18_-_8312848 | 1.13 |
ENSDART00000092033
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr17_-_3986236 | 1.12 |
ENSDART00000188794
ENSDART00000160830 |
PLCB1
|
si:ch1073-140o9.2 |
| chr2_+_50862527 | 1.10 |
ENSDART00000169800
ENSDART00000158847 ENSDART00000160900 |
adcyap1r1a
|
adenylate cyclase activating polypeptide 1a (pituitary) receptor type I |
| chr1_-_40227166 | 1.09 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr20_+_41664350 | 1.09 |
ENSDART00000186247
|
fam184a
|
family with sequence similarity 184, member A |
| chr11_-_36963988 | 1.09 |
ENSDART00000168288
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr7_-_5487593 | 1.09 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr5_+_61556172 | 1.08 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr13_-_7031033 | 1.07 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr5_-_69482891 | 1.07 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr2_+_20430366 | 1.06 |
ENSDART00000155108
|
si:ch211-153l6.6
|
si:ch211-153l6.6 |
| chr21_+_26612777 | 1.06 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr19_-_36675023 | 1.06 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr1_+_49266886 | 1.06 |
ENSDART00000137179
|
caly
|
calcyon neuron-specific vesicular protein |
| chr9_+_13682133 | 1.05 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr21_-_22485847 | 1.05 |
ENSDART00000162860
|
myo5b
|
myosin VB |
| chr15_+_15314189 | 1.05 |
ENSDART00000156830
|
camkk1b
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha b |
| chr10_-_20481763 | 1.05 |
ENSDART00000091156
ENSDART00000192358 |
tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr10_+_1638876 | 1.04 |
ENSDART00000184484
ENSDART00000060946 ENSDART00000181251 |
sgsm1b
|
small G protein signaling modulator 1b |
| chr3_+_56905103 | 1.04 |
ENSDART00000124728
ENSDART00000154408 |
otop2
|
otopetrin 2 |
| chr3_+_24986145 | 1.04 |
ENSDART00000055428
|
cbx7a
|
chromobox homolog 7a |
| chr17_-_45247151 | 1.02 |
ENSDART00000186230
|
ttbk2a
|
tau tubulin kinase 2a |
| chr18_-_12957451 | 1.02 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr19_+_4139065 | 1.02 |
ENSDART00000172524
|
si:dkey-218f9.10
|
si:dkey-218f9.10 |
| chr24_-_33366188 | 1.01 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr1_+_45323400 | 1.01 |
ENSDART00000148906
ENSDART00000132366 |
emp1
|
epithelial membrane protein 1 |
| chr2_+_47582488 | 1.00 |
ENSDART00000149967
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr7_+_25033924 | 1.00 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr13_+_13770980 | 0.99 |
ENSDART00000113089
|
slc4a11
|
solute carrier family 4, sodium borate transporter, member 11 |
| chr20_-_40750953 | 0.99 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr12_+_46967789 | 0.97 |
ENSDART00000114866
|
oat
|
ornithine aminotransferase |
| chr19_-_36234185 | 0.97 |
ENSDART00000186003
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr13_-_16222388 | 0.97 |
ENSDART00000182861
|
zgc:110045
|
zgc:110045 |
| chr14_+_49395437 | 0.97 |
ENSDART00000172850
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr20_-_39273505 | 0.96 |
ENSDART00000153114
|
clu
|
clusterin |
| chr17_+_41302660 | 0.96 |
ENSDART00000059480
|
fosl2
|
fos-like antigen 2 |
| chr3_+_39922602 | 0.96 |
ENSDART00000193937
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr5_+_4338874 | 0.95 |
ENSDART00000141866
|
sat1a.1
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 1 |
| chr3_+_7617353 | 0.95 |
ENSDART00000165551
|
zgc:109949
|
zgc:109949 |
| chr8_+_24861264 | 0.95 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr4_+_60492313 | 0.93 |
ENSDART00000191043
ENSDART00000191631 |
si:dkey-211i20.2
|
si:dkey-211i20.2 |
| chr5_+_20147830 | 0.93 |
ENSDART00000098727
|
svopa
|
SV2 related protein a |
| chr25_+_13498188 | 0.93 |
ENSDART00000015710
|
snrkb
|
SNF related kinase b |
| chr20_-_34090740 | 0.93 |
ENSDART00000062539
ENSDART00000008140 |
pdcb
|
phosducin b |
| chr7_+_17992455 | 0.93 |
ENSDART00000089211
ENSDART00000190881 |
ehbp1l1a
|
EH domain binding protein 1-like 1a |
| chr18_-_20869175 | 0.93 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr8_+_50983551 | 0.93 |
ENSDART00000142061
|
si:dkey-32e23.4
|
si:dkey-32e23.4 |
| chr8_+_24854600 | 0.92 |
ENSDART00000156570
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr6_+_59642695 | 0.91 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr23_-_12014931 | 0.91 |
ENSDART00000134652
|
si:dkey-178k16.1
|
si:dkey-178k16.1 |
| chr20_-_27864964 | 0.91 |
ENSDART00000153311
|
syndig1l
|
synapse differentiation inducing 1-like |
| chr1_-_34750169 | 0.90 |
ENSDART00000149380
|
si:dkey-151m22.8
|
si:dkey-151m22.8 |
| chr8_+_17933475 | 0.90 |
ENSDART00000100651
|
erich3
|
glutamate-rich 3 |
| chr12_-_43982343 | 0.90 |
ENSDART00000161539
|
CR385054.1
|
|
| chr3_+_4266289 | 0.90 |
ENSDART00000101636
|
si:dkey-73p2.1
|
si:dkey-73p2.1 |
| chr2_-_23778180 | 0.89 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr22_+_19188809 | 0.89 |
ENSDART00000134791
ENSDART00000133682 |
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr24_+_24923166 | 0.88 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr4_-_5291256 | 0.88 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr6_+_11250316 | 0.87 |
ENSDART00000137122
|
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr7_-_9674073 | 0.87 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr16_-_13613475 | 0.87 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr19_+_551963 | 0.87 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr19_+_43314700 | 0.87 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr19_+_5072918 | 0.86 |
ENSDART00000037126
|
eno2
|
enolase 2 |
| chr5_+_20035284 | 0.86 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr1_-_31505144 | 0.86 |
ENSDART00000087115
|
rims1b
|
regulating synaptic membrane exocytosis 1b |
| chr25_-_22191733 | 0.85 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr19_-_103289 | 0.85 |
ENSDART00000143118
|
adgrb1b
|
adhesion G protein-coupled receptor B1b |
| chr13_-_40141630 | 0.85 |
ENSDART00000181618
ENSDART00000171583 ENSDART00000192780 |
crtac1b
|
cartilage acidic protein 1b |
| chr16_+_48714048 | 0.84 |
ENSDART00000148709
ENSDART00000150121 |
brd2b
|
bromodomain containing 2b |
| chr9_-_21936841 | 0.84 |
ENSDART00000144843
|
lmo7a
|
LIM domain 7a |
| chr18_+_9635178 | 0.84 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr5_+_19337108 | 0.84 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr1_+_51312752 | 0.84 |
ENSDART00000063938
|
mast1a
|
microtubule associated serine/threonine kinase 1a |
| chr25_-_21085661 | 0.83 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr4_+_77735212 | 0.83 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr9_+_38481780 | 0.83 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr25_+_22643954 | 0.83 |
ENSDART00000182843
ENSDART00000121791 |
ush1c
|
Usher syndrome 1C |
| chr22_+_8979955 | 0.83 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr19_+_49721 | 0.82 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr17_+_8183393 | 0.82 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr12_-_10315039 | 0.82 |
ENSDART00000152680
|
pyyb
|
peptide YYb |
| chr6_+_27275716 | 0.81 |
ENSDART00000156434
ENSDART00000114347 |
scly
|
selenocysteine lyase |
| chr8_-_11170114 | 0.81 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr14_-_1535430 | 0.81 |
ENSDART00000179228
|
CABZ01109481.1
|
|
| chr2_+_30379650 | 0.80 |
ENSDART00000129542
|
crispld1b
|
cysteine-rich secretory protein LCCL domain containing 1b |
| chr9_-_22057658 | 0.80 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr20_-_16548912 | 0.80 |
ENSDART00000137601
|
ches1
|
checkpoint suppressor 1 |
| chr13_-_37545487 | 0.80 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr5_-_67629263 | 0.80 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_+_4573696 | 0.80 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr20_+_7084154 | 0.79 |
ENSDART00000136448
|
ftr85
|
finTRIM family, member 85 |
| chr16_+_13818500 | 0.79 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr12_-_4388704 | 0.79 |
ENSDART00000152168
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr2_+_47582681 | 0.79 |
ENSDART00000187579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr6_-_35446110 | 0.79 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr4_-_69189894 | 0.78 |
ENSDART00000169596
|
si:ch211-209j12.1
|
si:ch211-209j12.1 |
| chr7_+_14005111 | 0.78 |
ENSDART00000187365
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr9_-_22076368 | 0.78 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr18_+_5137241 | 0.77 |
ENSDART00000159601
|
serpini1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr15_-_43978141 | 0.77 |
ENSDART00000041249
|
chordc1a
|
cysteine and histidine-rich domain (CHORD) containing 1a |
| chr9_-_12034444 | 0.77 |
ENSDART00000038651
|
znf804a
|
zinc finger protein 804A |
| chr23_-_7797207 | 0.76 |
ENSDART00000181611
|
myt1b
|
myelin transcription factor 1b |
| chr4_+_76671012 | 0.76 |
ENSDART00000005585
|
ms4a17a.2
|
membrane-spanning 4-domains, subfamily A, member 17a.2 |
| chr23_-_18024543 | 0.76 |
ENSDART00000139695
|
pm20d1.1
|
peptidase M20 domain containing 1, tandem duplicate 1 |
| chr8_+_44714336 | 0.76 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr20_+_34915945 | 0.76 |
ENSDART00000153064
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr8_+_14792830 | 0.76 |
ENSDART00000139972
|
cacna1ea
|
calcium channel, voltage-dependent, R type, alpha 1E subunit a |
| chr5_+_15350954 | 0.75 |
ENSDART00000140990
ENSDART00000137287 ENSDART00000061653 |
pebp1
|
phosphatidylethanolamine binding protein 1 |
| chr17_-_29192987 | 0.75 |
ENSDART00000164302
|
sptbn5
|
spectrin, beta, non-erythrocytic 5 |
| chr3_+_49097775 | 0.74 |
ENSDART00000169185
|
zgc:123284
|
zgc:123284 |
| chr13_-_10945288 | 0.74 |
ENSDART00000114315
ENSDART00000164667 ENSDART00000159482 |
abcg8
|
ATP-binding cassette, sub-family G (WHITE), member 8 |
| chr2_-_56387041 | 0.74 |
ENSDART00000036240
|
cers4b
|
ceramide synthase 4b |
| chr5_-_35159379 | 0.74 |
ENSDART00000144105
|
fcho2
|
FCH domain only 2 |
| chr17_-_12389259 | 0.74 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr17_-_45247332 | 0.73 |
ENSDART00000016815
|
ttbk2a
|
tau tubulin kinase 2a |
| chr4_+_76752707 | 0.73 |
ENSDART00000082121
|
ms4a17a.4
|
membrane-spanning 4-domains, subfamily A, member 17A.4 |
| chr3_+_45401472 | 0.72 |
ENSDART00000156693
|
baiap3
|
BAI1-associated protein 3 |
| chr24_+_39135419 | 0.72 |
ENSDART00000180941
|
tbc1d24
|
TBC1 domain family, member 24 |
| chr9_-_30363770 | 0.72 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr2_+_31600661 | 0.71 |
ENSDART00000139039
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr19_-_5805923 | 0.71 |
ENSDART00000134340
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr19_-_22621991 | 0.71 |
ENSDART00000175106
|
pleca
|
plectin a |
| chr24_+_26039464 | 0.71 |
ENSDART00000131017
|
tnk2a
|
tyrosine kinase, non-receptor, 2a |
| chr8_-_38403018 | 0.71 |
ENSDART00000134100
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr2_+_3452590 | 0.71 |
ENSDART00000153935
|
cyp8b2
|
cytochrome P450, family 8, subfamily B, polypeptide 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smarcc1a+smarcc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.6 | 1.9 | GO:0015824 | proline transport(GO:0015824) |
| 0.5 | 2.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.5 | 1.4 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.4 | 1.6 | GO:0048913 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.3 | 1.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.3 | 1.3 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.3 | 1.1 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.3 | 2.3 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 2.6 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.2 | 1.0 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.2 | 0.6 | GO:0099538 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.2 | 0.8 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 0.8 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.7 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.2 | 0.6 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.8 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.2 | 0.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.2 | 2.4 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.2 | 0.5 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.2 | 0.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 | 0.5 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.4 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 1.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.7 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.8 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 1.6 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.1 | 1.0 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 2.4 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.4 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 1.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.5 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.1 | 0.4 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 1.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 0.9 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.3 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.9 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.6 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 1.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.6 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 0.4 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 1.3 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.1 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 1.0 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 1.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.5 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.1 | 0.2 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.1 | GO:0060347 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 2.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.1 | 1.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 1.1 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.1 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 0.4 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.1 | 0.3 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.4 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 1.1 | GO:0051496 | positive regulation of stress fiber assembly(GO:0051496) |
| 0.1 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 0.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.3 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.5 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.2 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 2.1 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.1 | 0.9 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.1 | 0.7 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.5 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 6.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.5 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.0 | 0.4 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 2.4 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 1.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.2 | GO:0070459 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.3 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.7 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.8 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.4 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.7 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.0 | 3.0 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 5.0 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 7.3 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 1.0 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 1.4 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 1.9 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.6 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.2 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.5 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) Schwann cell migration(GO:0036135) cardiac neuron differentiation(GO:0060945) cardiac neuron development(GO:0060959) |
| 0.0 | 0.7 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0090134 | coronary vasculature development(GO:0060976) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.3 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 1.6 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.2 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 1.2 | GO:0046328 | regulation of JNK cascade(GO:0046328) |
| 0.0 | 0.5 | GO:0007040 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 2.3 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 2.6 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.8 | GO:0051298 | centrosome duplication(GO:0051298) |
| 0.0 | 0.5 | GO:0050868 | negative regulation of T cell activation(GO:0050868) negative regulation of leukocyte cell-cell adhesion(GO:1903038) |
| 0.0 | 0.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 1.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.6 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.8 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.9 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.5 | GO:0097553 | release of sequestered calcium ion into cytosol(GO:0051209) calcium ion transmembrane import into cytosol(GO:0097553) calcium ion import into cytosol(GO:1902656) |
| 0.0 | 0.3 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 1.0 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:1901071 | glucosamine-containing compound metabolic process(GO:1901071) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0043247 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.4 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0050708 | regulation of protein secretion(GO:0050708) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.8 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.1 | GO:0099565 | excitatory postsynaptic potential(GO:0060079) chemical synaptic transmission, postsynaptic(GO:0099565) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.2 | 1.5 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 0.5 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.2 | 1.3 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 1.5 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 2.8 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 2.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.8 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.1 | 0.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.7 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.5 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.5 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.5 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 0.3 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 0.4 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 2.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 2.1 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.0 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.8 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 2.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.9 | GO:0098831 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 3.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 3.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.9 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 2.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.5 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.5 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.1 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.9 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0008397 | sterol 12-alpha-hydroxylase activity(GO:0008397) |
| 0.6 | 3.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.3 | 2.3 | GO:0071916 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.3 | 2.4 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 2.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 0.7 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 1.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) neuregulin binding(GO:0038132) |
| 0.2 | 0.6 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.6 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.2 | 0.6 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 1.7 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.2 | 1.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.2 | 0.9 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 2.6 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 0.7 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.2 | 0.7 | GO:0072570 | ADP-D-ribose binding(GO:0072570) mono-ADP-D-ribose binding(GO:0072571) |
| 0.2 | 0.8 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.2 | 1.3 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.2 | 3.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 2.1 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.0 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 0.2 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 1.0 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 1.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.8 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 3.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 3.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 7.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0004133 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.1 | 0.5 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.4 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.4 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.5 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.3 | GO:0043121 | neurotrophin binding(GO:0043121) |
| 0.1 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.7 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.6 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 0.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 0.2 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 5.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.9 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.8 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 1.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.4 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 1.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 6.0 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 1.0 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.2 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:1900750 | oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 6.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.2 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 3.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 1.5 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.5 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 1.0 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.3 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.6 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.7 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 2.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 5.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.5 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.4 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.1 | 3.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 0.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 1.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.2 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 1.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.7 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.6 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.3 | 0.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 2.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.2 | 0.5 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 1.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.7 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.5 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 2.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.5 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.6 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.7 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.7 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.9 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |