Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for smad2_smad10a+smad4a_smad1
Z-value: 1.36
Transcription factors associated with smad2_smad10a+smad4a_smad1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
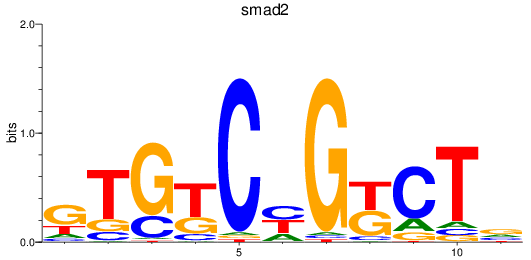
smad2
|
ENSDARG00000006389 | SMAD family member 2 |
|
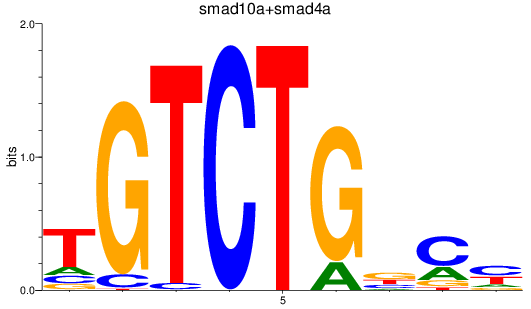
smad10a
|
ENSDARG00000045094 | si_dkey-222b8.1 |
|
smad10a
|
ENSDARG00000070428 | si_dkey-222b8.1 |
|
smad4a
|
ENSDARG00000075226 | SMAD family member 4a |
|
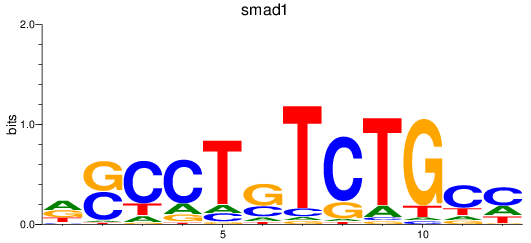
smad1
|
ENSDARG00000027199 | SMAD family member 1 |
|
smad1
|
ENSDARG00000112617 | SMAD family member 1 |
|
smad1
|
ENSDARG00000115674 | SMAD family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad2 | dr11_v1_chr10_-_14920989_14920989 | 0.79 | 6.1e-05 | Click! |
| si:dkey-239n17.4 | dr11_v1_chr16_-_28876479_28876479 | 0.49 | 3.1e-02 | Click! |
| si:dkey-222b8.1 | dr11_v1_chr19_+_23932259_23932259 | 0.43 | 6.9e-02 | Click! |
| smad4a | dr11_v1_chr10_+_585719_585719 | 0.29 | 2.3e-01 | Click! |
| smad1 | dr11_v1_chr1_-_35916247_35916247 | 0.26 | 2.9e-01 | Click! |
Activity profile of smad2_smad10a+smad4a_smad1 motif
Sorted Z-values of smad2_smad10a+smad4a_smad1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_46040618 | 4.73 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr13_-_479129 | 2.72 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr20_-_7648325 | 2.25 |
ENSDART00000186541
|
CR848049.1
|
|
| chr23_+_36460239 | 2.17 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr9_-_180334 | 1.97 |
ENSDART00000180339
|
zgc:158619
|
zgc:158619 |
| chr2_+_54755172 | 1.89 |
ENSDART00000097864
|
ankrd12
|
ankyrin repeat domain 12 |
| chr3_-_3496738 | 1.84 |
ENSDART00000186849
|
CABZ01040998.1
|
|
| chr14_+_8127893 | 1.73 |
ENSDART00000169091
|
psd2
|
pleckstrin and Sec7 domain containing 2 |
| chr5_-_66028714 | 1.73 |
ENSDART00000022625
ENSDART00000164228 |
nrarpb
|
NOTCH regulated ankyrin repeat protein b |
| chr13_-_5079511 | 1.59 |
ENSDART00000110248
|
slc29a3
|
solute carrier family 29 (equilibrative nucleoside transporter), member 3 |
| chr25_+_31958911 | 1.56 |
ENSDART00000191394
ENSDART00000090727 ENSDART00000185893 |
duox
|
dual oxidase |
| chr22_+_635813 | 1.50 |
ENSDART00000179067
|
CU856139.1
|
|
| chr18_+_50461981 | 1.44 |
ENSDART00000158761
|
CU896640.1
|
|
| chr10_-_10018794 | 1.26 |
ENSDART00000130734
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr14_-_237130 | 1.25 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr8_+_32719930 | 1.17 |
ENSDART00000145362
|
hmcn2
|
hemicentin 2 |
| chr20_+_35085224 | 1.14 |
ENSDART00000040456
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr23_+_41831224 | 1.10 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr6_-_31827597 | 1.07 |
ENSDART00000159400
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr4_-_76488581 | 1.05 |
ENSDART00000174291
|
ftr51
|
finTRIM family, member 51 |
| chr7_-_72269049 | 1.04 |
ENSDART00000161497
|
CU463038.2
|
|
| chr14_+_8442248 | 1.01 |
ENSDART00000163059
|
si:dkey-160o24.3
|
si:dkey-160o24.3 |
| chr17_+_1514711 | 0.99 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr19_+_41551335 | 0.99 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr13_+_25433774 | 0.96 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr10_+_37181780 | 0.95 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr19_+_42219165 | 0.93 |
ENSDART00000163192
|
CU896644.1
|
|
| chr9_+_45605410 | 0.86 |
ENSDART00000136444
ENSDART00000007189 ENSDART00000158713 ENSDART00000182671 |
traf3ip1
|
TNF receptor-associated factor 3 interacting protein 1 |
| chr9_+_52411530 | 0.86 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr20_-_48061351 | 0.80 |
ENSDART00000164962
|
prep
|
prolyl endopeptidase |
| chr12_-_9294819 | 0.80 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr5_-_58780160 | 0.75 |
ENSDART00000166955
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr19_+_31061718 | 0.75 |
ENSDART00000145971
|
sostdc1b
|
sclerostin domain containing 1b |
| chr18_+_49225552 | 0.72 |
ENSDART00000135026
|
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr5_+_18047111 | 0.70 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr15_+_28303161 | 0.70 |
ENSDART00000087926
|
myo1cb
|
myosin Ic, paralog b |
| chr2_-_42628028 | 0.68 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr18_-_44908479 | 0.67 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr23_+_44080382 | 0.67 |
ENSDART00000011715
|
zgc:56304
|
zgc:56304 |
| chr4_-_16412084 | 0.66 |
ENSDART00000188460
|
dcn
|
decorin |
| chr23_+_40908583 | 0.64 |
ENSDART00000180933
|
LO017845.1
|
|
| chr19_+_25478203 | 0.64 |
ENSDART00000132934
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr17_+_45765164 | 0.62 |
ENSDART00000145882
|
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr17_+_50524573 | 0.60 |
ENSDART00000187974
|
CR382385.1
|
|
| chr11_+_77526 | 0.59 |
ENSDART00000193521
|
CABZ01072242.1
|
|
| chr24_-_26632171 | 0.57 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr16_-_39859119 | 0.54 |
ENSDART00000138388
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr22_-_506522 | 0.54 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr3_-_1317290 | 0.53 |
ENSDART00000047094
|
LO018552.1
|
|
| chr20_+_18163355 | 0.52 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr25_+_32157326 | 0.50 |
ENSDART00000112588
|
tjp1b
|
tight junction protein 1b |
| chr13_+_22295905 | 0.49 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr16_-_11961325 | 0.48 |
ENSDART00000143845
|
si:ch73-296e2.3
|
si:ch73-296e2.3 |
| chr8_+_28527776 | 0.48 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| chr2_+_47623202 | 0.48 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr7_-_18508815 | 0.47 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr25_-_6261693 | 0.46 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr19_-_18152942 | 0.44 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr6_+_6491013 | 0.43 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr4_-_76488854 | 0.42 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr5_+_457729 | 0.42 |
ENSDART00000025183
|
ift74
|
intraflagellar transport 74 |
| chr3_+_13879446 | 0.41 |
ENSDART00000164767
|
farsa
|
phenylalanyl-tRNA synthetase, alpha subunit |
| chr20_-_26042070 | 0.41 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
| chr19_-_47456787 | 0.41 |
ENSDART00000168792
|
tfap2e
|
transcription factor AP-2 epsilon |
| chr1_+_57347888 | 0.41 |
ENSDART00000104222
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr18_-_5542351 | 0.41 |
ENSDART00000129459
|
spata5l1
|
spermatogenesis associated 5-like 1 |
| chr8_-_2230128 | 0.41 |
ENSDART00000140427
|
si:dkeyp-117b11.2
|
si:dkeyp-117b11.2 |
| chr11_+_45255774 | 0.40 |
ENSDART00000172838
|
aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr5_-_70625697 | 0.40 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr25_-_210730 | 0.40 |
ENSDART00000187580
|
FP236318.1
|
|
| chr4_-_191736 | 0.40 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr18_+_50961953 | 0.40 |
ENSDART00000158768
|
ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chr6_-_52757276 | 0.39 |
ENSDART00000159154
|
AL954861.2
|
|
| chr7_+_26998169 | 0.38 |
ENSDART00000128110
ENSDART00000101018 |
caprin1a
|
cell cycle associated protein 1a |
| chr3_-_31019715 | 0.37 |
ENSDART00000156687
ENSDART00000156127 ENSDART00000153945 |
si:dkeyp-71f10.5
|
si:dkeyp-71f10.5 |
| chr9_+_51882534 | 0.37 |
ENSDART00000165493
|
CABZ01079550.1
|
|
| chr15_-_40157331 | 0.36 |
ENSDART00000187958
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr7_-_29271738 | 0.35 |
ENSDART00000109030
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr19_+_41551543 | 0.35 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr16_-_30655980 | 0.34 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr20_-_2355357 | 0.34 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr3_-_53559408 | 0.34 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr20_-_4049862 | 0.33 |
ENSDART00000158057
|
sprtn
|
SprT-like N-terminal domain |
| chr4_-_21466825 | 0.33 |
ENSDART00000066897
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr23_-_44965582 | 0.33 |
ENSDART00000163367
|
tfr2
|
transferrin receptor 2 |
| chr8_+_54055390 | 0.33 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr10_-_35186310 | 0.33 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr22_+_35930526 | 0.32 |
ENSDART00000169242
|
LO016987.1
|
|
| chr13_-_16226312 | 0.32 |
ENSDART00000163952
|
zgc:110045
|
zgc:110045 |
| chr6_+_11989537 | 0.32 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr14_-_451555 | 0.32 |
ENSDART00000190906
|
FAT4
|
FAT atypical cadherin 4 |
| chr19_-_36675023 | 0.32 |
ENSDART00000132471
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr22_+_38824012 | 0.32 |
ENSDART00000144318
ENSDART00000003736 |
anos1b
|
anosmin 1b |
| chr14_+_6546516 | 0.32 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr20_+_23625387 | 0.32 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr19_-_43819582 | 0.31 |
ENSDART00000160879
ENSDART00000075902 |
klhl43
|
kelch-like family member 43 |
| chr3_-_53559581 | 0.30 |
ENSDART00000183499
|
notch3
|
notch 3 |
| chr12_-_628024 | 0.30 |
ENSDART00000158563
ENSDART00000152612 |
wu:fj29h11
si:ch73-301j1.1
|
wu:fj29h11 si:ch73-301j1.1 |
| chr20_+_52442870 | 0.30 |
ENSDART00000163100
|
arhgap39
|
Rho GTPase activating protein 39 |
| chr12_+_48581588 | 0.30 |
ENSDART00000114996
|
CABZ01078415.1
|
|
| chr23_-_27152866 | 0.30 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr15_-_40157513 | 0.29 |
ENSDART00000184014
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr1_+_47446032 | 0.29 |
ENSDART00000126904
ENSDART00000007262 |
gja8b
|
gap junction protein alpha 8 paralog b |
| chr5_-_66749535 | 0.29 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr1_+_47585700 | 0.29 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr12_+_17933775 | 0.29 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr19_+_4061699 | 0.29 |
ENSDART00000158309
ENSDART00000166512 |
btr25
btr26
|
bloodthirsty-related gene family, member 25 bloodthirsty-related gene family, member 26 |
| chr17_+_10578823 | 0.29 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr9_+_34148714 | 0.29 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr19_-_13286722 | 0.28 |
ENSDART00000168296
ENSDART00000158330 |
zfpm2b
|
zinc finger protein, FOG family member 2b |
| chr25_+_32496723 | 0.28 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr4_+_41602 | 0.28 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr20_+_54356540 | 0.28 |
ENSDART00000143591
|
znf410
|
zinc finger protein 410 |
| chr18_+_50924556 | 0.28 |
ENSDART00000159287
|
pik3c2g
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 gamma |
| chr13_+_12175724 | 0.28 |
ENSDART00000166053
|
gabrg1
|
gamma-aminobutyric acid type A receptor gamma1 subunit |
| chr15_+_45595385 | 0.28 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr21_-_45728069 | 0.28 |
ENSDART00000185101
|
CU302316.1
|
|
| chr1_-_51157660 | 0.27 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr9_-_22963897 | 0.27 |
ENSDART00000133676
|
si:dkey-91i10.2
|
si:dkey-91i10.2 |
| chr23_+_45734011 | 0.27 |
ENSDART00000062415
|
CABZ01088025.1
|
|
| chr12_+_33403694 | 0.27 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr19_-_38611814 | 0.27 |
ENSDART00000151958
|
col16a1
|
collagen, type XVI, alpha 1 |
| chr24_+_38306010 | 0.27 |
ENSDART00000143184
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr2_-_42705284 | 0.27 |
ENSDART00000187160
|
myo10
|
myosin X |
| chr11_-_44999858 | 0.27 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr17_+_6467398 | 0.27 |
ENSDART00000149527
|
COQ8A (1 of many)
|
si:dkey-36g24.3 |
| chr17_-_608857 | 0.27 |
ENSDART00000163431
|
KLHL28
|
kelch like family member 28 |
| chr5_-_41638039 | 0.27 |
ENSDART00000144525
|
epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr14_-_22390821 | 0.27 |
ENSDART00000054396
|
fbxl3l
|
F-box and leucine-rich repeat protein 3, like |
| chr23_+_4362463 | 0.26 |
ENSDART00000039829
|
zgc:112175
|
zgc:112175 |
| chr12_+_30789611 | 0.26 |
ENSDART00000181501
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr1_-_55238610 | 0.26 |
ENSDART00000110818
|
FO704915.1
|
|
| chr18_+_27337994 | 0.26 |
ENSDART00000136172
|
si:dkey-29p10.4
|
si:dkey-29p10.4 |
| chr18_+_5549672 | 0.26 |
ENSDART00000184970
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr1_-_54765262 | 0.26 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr5_+_483965 | 0.26 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr19_-_3754327 | 0.25 |
ENSDART00000168463
|
btr21
|
bloodthirsty-related gene family, member 21 |
| chr25_+_32496877 | 0.25 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr12_+_48511605 | 0.25 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr7_+_16352924 | 0.25 |
ENSDART00000158972
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr7_-_25895189 | 0.25 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr14_-_1949277 | 0.25 |
ENSDART00000159435
|
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr1_-_44037843 | 0.24 |
ENSDART00000160276
|
si:ch73-109d9.3
|
si:ch73-109d9.3 |
| chr13_+_45980163 | 0.24 |
ENSDART00000074547
ENSDART00000005195 |
bsdc1
|
BSD domain containing 1 |
| chr24_+_41610068 | 0.24 |
ENSDART00000159507
|
CABZ01044108.1
|
|
| chr10_+_37182626 | 0.24 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr4_-_29753895 | 0.24 |
ENSDART00000150742
|
si:dkey-191j3.1
|
si:dkey-191j3.1 |
| chr17_+_24843401 | 0.24 |
ENSDART00000110179
|
cx34.4
|
connexin 34.4 |
| chr5_+_36752943 | 0.24 |
ENSDART00000017138
|
exoc3l2a
|
exocyst complex component 3-like 2a |
| chr1_-_59411901 | 0.24 |
ENSDART00000167015
|
si:ch211-188p14.4
|
si:ch211-188p14.4 |
| chr13_+_38302665 | 0.24 |
ENSDART00000145777
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr6_+_9793495 | 0.24 |
ENSDART00000108524
|
als2b
|
amyotrophic lateral sclerosis 2b (juvenile) |
| chr13_-_36418921 | 0.24 |
ENSDART00000135804
|
dcaf5
|
ddb1 and cul4 associated factor 5 |
| chr4_+_77740228 | 0.24 |
ENSDART00000193397
|
si:zfos-2131b9.2
|
si:zfos-2131b9.2 |
| chr25_+_37268900 | 0.24 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr2_+_26179096 | 0.23 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr3_-_32320537 | 0.23 |
ENSDART00000113550
ENSDART00000168483 |
si:dkey-16p21.7
|
si:dkey-16p21.7 |
| chr24_-_37272116 | 0.23 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr10_+_42589391 | 0.23 |
ENSDART00000067689
ENSDART00000075259 |
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr12_+_45060789 | 0.23 |
ENSDART00000190396
|
ADAM12 (1 of many)
|
ADAM metallopeptidase domain 12 |
| chr21_+_10828828 | 0.23 |
ENSDART00000136449
|
oacyl
|
O-acyltransferase like |
| chr7_+_36898622 | 0.23 |
ENSDART00000190773
|
tox3
|
TOX high mobility group box family member 3 |
| chr19_+_25497541 | 0.23 |
ENSDART00000144300
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr25_-_31433512 | 0.22 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr10_-_74408 | 0.22 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr21_+_26390549 | 0.22 |
ENSDART00000185643
|
tmsb
|
thymosin, beta |
| chr14_-_30050 | 0.22 |
ENSDART00000164411
|
zbtb49
|
zinc finger and BTB domain containing 49 |
| chr23_+_3594171 | 0.22 |
ENSDART00000159609
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr5_+_8964926 | 0.22 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr23_+_44633858 | 0.22 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr2_+_19236677 | 0.22 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr3_-_21288202 | 0.21 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr18_+_6054816 | 0.21 |
ENSDART00000113668
|
si:ch73-386h18.1
|
si:ch73-386h18.1 |
| chr4_+_4889257 | 0.21 |
ENSDART00000175651
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr20_+_50061890 | 0.21 |
ENSDART00000137725
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr21_-_41588129 | 0.21 |
ENSDART00000164125
|
crebrf
|
creb3 regulatory factor |
| chr10_+_44581378 | 0.21 |
ENSDART00000190331
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr19_-_81851 | 0.21 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_+_24458899 | 0.21 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr3_-_38783951 | 0.20 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr18_+_18612388 | 0.20 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr4_+_31259 | 0.20 |
ENSDART00000166826
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr10_+_1052591 | 0.20 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr4_+_11458078 | 0.20 |
ENSDART00000037600
|
ankrd16
|
ankyrin repeat domain 16 |
| chr4_+_76957765 | 0.20 |
ENSDART00000129607
|
si:dkey-240n22.6
|
si:dkey-240n22.6 |
| chr6_-_55958705 | 0.20 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr19_-_42503143 | 0.20 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr21_+_10866421 | 0.20 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr25_-_31396479 | 0.20 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr4_-_25858620 | 0.20 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr19_+_42754882 | 0.20 |
ENSDART00000133711
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr18_+_6706140 | 0.20 |
ENSDART00000111343
|
lmf2a
|
lipase maturation factor 2a |
| chr23_-_874418 | 0.20 |
ENSDART00000133906
|
rbm10
|
RNA binding motif protein 10 |
| chr19_-_48010490 | 0.20 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr10_-_300000 | 0.20 |
ENSDART00000183273
|
emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr20_-_1265562 | 0.20 |
ENSDART00000189866
|
lats1
|
large tumor suppressor kinase 1 |
| chr10_-_44341288 | 0.20 |
ENSDART00000166131
|
zswim6
|
zinc finger, SWIM-type containing 6 |
| chr20_+_44582318 | 0.19 |
ENSDART00000149000
ENSDART00000149775 ENSDART00000085416 |
atad2b
|
ATPase family, AAA domain containing 2B |
| chr23_+_3602779 | 0.19 |
ENSDART00000013629
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr9_+_12907574 | 0.19 |
ENSDART00000102348
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad2_smad10a+smad4a_smad1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.2 | 1.1 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.2 | 0.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.2 | 0.6 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 1.2 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.2 | 1.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 1.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.4 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.1 | 0.9 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) cerebrospinal fluid circulation(GO:0090660) |
| 0.1 | 1.6 | GO:1901642 | nucleoside transport(GO:0015858) nucleoside transmembrane transport(GO:1901642) |
| 0.1 | 0.4 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.1 | 0.6 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.4 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.1 | 1.7 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.9 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.7 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 0.2 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.5 | GO:0006833 | water transport(GO:0006833) |
| 0.1 | 0.3 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.3 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.2 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 1.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 0.3 | GO:0032616 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.3 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 2.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.8 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.0 | 0.2 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.1 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.5 | GO:0043516 | regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043516) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.2 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.0 | 0.2 | GO:1902837 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.3 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.0 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.1 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.0 | 0.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.5 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.1 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.2 | GO:0034333 | adherens junction assembly(GO:0034333) |
| 0.0 | 1.5 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.2 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.9 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.0 | 2.1 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.2 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.1 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.0 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 1.1 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.5 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.0 | 0.8 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.1 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.0 | 0.2 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.8 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0090311 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.0 | 0.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:0008584 | male gonad development(GO:0008584) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.3 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.5 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.1 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.0 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.3 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.0 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.4 | GO:0048920 | neuromast primordium migration(GO:0048883) posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 0.3 | GO:0035675 | neuromast hair cell development(GO:0035675) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.2 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.1 | GO:0001765 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) |
| 0.0 | 0.2 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.1 | GO:0048886 | neuromast hair cell differentiation(GO:0048886) |
| 0.0 | 0.1 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0042044 | fluid transport(GO:0042044) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.3 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.0 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.0 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.0 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0009648 | photoperiodism(GO:0009648) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 2.9 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 0.1 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.0 | 0.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 1.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.3 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.2 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.0 | GO:0098753 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 0.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.0 | GO:0005913 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 0.5 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.4 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 1.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.6 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.3 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 0.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.2 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.2 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0032138 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 1.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 1.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.3 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 2.1 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.0 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.5 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.6 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.1 | 0.7 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.2 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 1.2 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.3 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.1 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |