Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
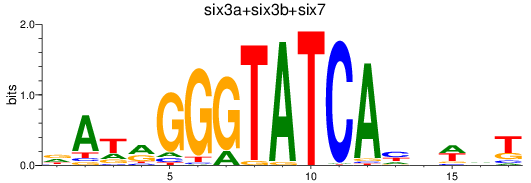
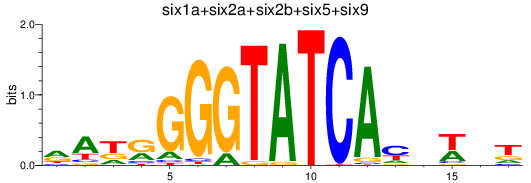
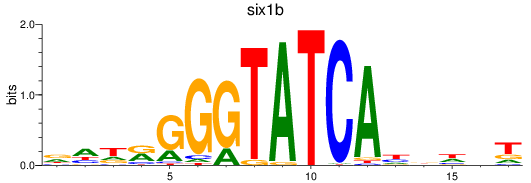
Results for six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
Z-value: 0.88
Transcription factors associated with six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six3b
|
ENSDARG00000054879 | SIX homeobox 3b |
|
six3a
|
ENSDARG00000058008 | SIX homeobox 3a |
|
six7
|
ENSDARG00000070107 | SIX homeobox 7 |
|
six3a
|
ENSDARG00000114971 | SIX homeobox 3a |
|
six1a
|
ENSDARG00000039304 | SIX homeobox 1a |
|
six2b
|
ENSDARG00000054878 | SIX homeobox 2b |
|
six2a
|
ENSDARG00000058004 | SIX homeobox 2a |
|
six5
|
ENSDARG00000068406 | SIX homeobox 5 |
|
six9
|
ENSDARG00000068407 | SIX homeobox 9 |
|
six2a
|
ENSDARG00000115214 | SIX homeobox 2a |
|
six1b
|
ENSDARG00000026473 | SIX homeobox 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six3b | dr11_v1_chr12_+_25600685_25600685 | -0.92 | 2.5e-08 | Click! |
| six1a | dr11_v1_chr13_-_31622195_31622195 | -0.75 | 2.4e-04 | Click! |
| six7 | dr11_v1_chr7_-_7420301_7420301 | 0.55 | 1.5e-02 | Click! |
| six9 | dr11_v1_chr18_-_37241080_37241115 | 0.45 | 5.5e-02 | Click! |
| six2b | dr11_v1_chr12_-_25612170_25612170 | -0.45 | 5.5e-02 | Click! |
| six2a | dr11_v1_chr13_+_10232695_10232695 | 0.38 | 1.1e-01 | Click! |
| six1b | dr11_v1_chr20_+_20499869_20499869 | 0.22 | 3.7e-01 | Click! |
| six5 | dr11_v1_chr18_-_37252036_37252036 | 0.09 | 7.1e-01 | Click! |
| six3a | dr11_v1_chr13_-_10261383_10261383 | -0.06 | 8.0e-01 | Click! |
Activity profile of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b motif
Sorted Z-values of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41181869 | 7.47 |
ENSDART00000002046
|
opn1mw1
|
opsin 1 (cone pigments), medium-wave-sensitive, 1 |
| chr17_+_20607487 | 4.17 |
ENSDART00000154123
|
si:ch73-288o11.5
|
si:ch73-288o11.5 |
| chr5_+_68112194 | 3.13 |
ENSDART00000162468
|
CABZ01083937.1
|
|
| chr25_-_19068557 | 2.84 |
ENSDART00000184780
|
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr23_-_39666519 | 2.73 |
ENSDART00000110868
ENSDART00000190961 |
vwa1
|
von Willebrand factor A domain containing 1 |
| chr1_-_24255149 | 2.65 |
ENSDART00000146960
|
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr11_+_24002503 | 2.31 |
ENSDART00000164702
|
chia.2
|
chitinase, acidic.2 |
| chr7_-_71486162 | 2.20 |
ENSDART00000045253
|
aco1
|
aconitase 1, soluble |
| chr5_+_9377005 | 2.15 |
ENSDART00000124924
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr1_-_53259746 | 2.12 |
ENSDART00000134222
|
il15
|
interleukin 15 |
| chr21_+_11385031 | 1.87 |
ENSDART00000040598
|
cel.2
|
carboxyl ester lipase, tandem duplicate 2 |
| chr4_-_27398385 | 1.72 |
ENSDART00000142117
ENSDART00000150553 ENSDART00000182746 |
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr11_+_3575963 | 1.71 |
ENSDART00000077305
|
si:dkey-33m11.8
|
si:dkey-33m11.8 |
| chr1_+_37752171 | 1.64 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr9_+_46644633 | 1.60 |
ENSDART00000160285
|
slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr13_-_47800342 | 1.53 |
ENSDART00000121817
|
fbln7
|
fibulin 7 |
| chr6_-_48087152 | 1.53 |
ENSDART00000180614
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr6_+_41186320 | 1.42 |
ENSDART00000025241
|
opn1mw2
|
opsin 1 (cone pigments), medium-wave-sensitive, 2 |
| chr20_-_34127415 | 1.39 |
ENSDART00000010028
|
ptgs2b
|
prostaglandin-endoperoxide synthase 2b |
| chr4_-_27301356 | 1.33 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr23_+_39481537 | 1.28 |
ENSDART00000187222
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr11_-_24016761 | 1.20 |
ENSDART00000153601
ENSDART00000067817 ENSDART00000170531 |
chia.3
|
chitinase, acidic.3 |
| chr19_+_34230108 | 1.17 |
ENSDART00000141950
|
galnt12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 |
| chr5_-_12823876 | 1.17 |
ENSDART00000019461
|
p2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr15_+_17446796 | 1.15 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr1_+_532766 | 1.10 |
ENSDART00000179731
ENSDART00000060944 |
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr13_-_40120252 | 1.06 |
ENSDART00000157852
|
crtac1b
|
cartilage acidic protein 1b |
| chr17_-_44776224 | 1.06 |
ENSDART00000156195
|
zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr1_+_7538909 | 1.00 |
ENSDART00000148276
|
mylz3
|
myosin, light polypeptide 3, skeletal muscle |
| chr7_-_47381146 | 0.97 |
ENSDART00000185596
|
AL935126.2
|
|
| chr5_-_30615901 | 0.96 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr10_+_23520952 | 0.93 |
ENSDART00000125317
|
zmp:0000000937
|
zmp:0000000937 |
| chr10_-_20706109 | 0.93 |
ENSDART00000124751
|
kcnip3b
|
Kv channel interacting protein 3b, calsenilin |
| chr4_+_14826299 | 0.92 |
ENSDART00000067035
|
shisal1a
|
shisa like 1a |
| chr12_+_13742778 | 0.91 |
ENSDART00000111401
ENSDART00000190552 |
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr12_+_13742953 | 0.89 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr9_-_22076368 | 0.87 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr7_-_38183331 | 0.85 |
ENSDART00000149382
|
abcc12
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 12 |
| chr20_-_45709990 | 0.79 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr15_-_36352983 | 0.77 |
ENSDART00000059786
|
si:dkey-23k10.5
|
si:dkey-23k10.5 |
| chr16_+_50755133 | 0.77 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr16_+_41060161 | 0.77 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr24_+_30435164 | 0.71 |
ENSDART00000193877
|
dpyda.1
|
dihydropyrimidine dehydrogenase a, tandem duplicate 1 |
| chr10_-_42898220 | 0.70 |
ENSDART00000099270
|
CU326366.2
|
|
| chr13_-_18122333 | 0.67 |
ENSDART00000128748
|
washc2c
|
WASH complex subunit 2C |
| chr11_-_44194132 | 0.67 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr14_+_26805388 | 0.66 |
ENSDART00000044389
ENSDART00000173126 |
klhl4
|
kelch-like family member 4 |
| chr8_+_23745167 | 0.66 |
ENSDART00000149352
|
si:ch211-163l21.11
|
si:ch211-163l21.11 |
| chr1_-_21901589 | 0.65 |
ENSDART00000140553
|
frmpd1a
|
FERM and PDZ domain containing 1a |
| chr5_+_64732036 | 0.65 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr21_+_18405585 | 0.63 |
ENSDART00000139318
|
si:dkey-1d7.3
|
si:dkey-1d7.3 |
| chr6_-_40899618 | 0.61 |
ENSDART00000153949
ENSDART00000021969 |
zgc:172271
|
zgc:172271 |
| chr20_+_22067337 | 0.58 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr13_+_9678427 | 0.58 |
ENSDART00000153907
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr7_-_72423666 | 0.58 |
ENSDART00000191214
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr3_-_5067585 | 0.52 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr13_-_2215213 | 0.51 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr25_+_192116 | 0.50 |
ENSDART00000153983
|
zgc:114188
|
zgc:114188 |
| chr16_-_383664 | 0.49 |
ENSDART00000051693
|
irx4a
|
iroquois homeobox 4a |
| chr10_+_21718468 | 0.47 |
ENSDART00000126622
|
pcdh1gb9
|
protocadherin 1 gamma b 9 |
| chr8_-_26961779 | 0.47 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr20_-_39735952 | 0.44 |
ENSDART00000101049
ENSDART00000137485 ENSDART00000062402 |
tpd52l1
|
tumor protein D52-like 1 |
| chr10_+_35468978 | 0.43 |
ENSDART00000131984
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr10_-_32610776 | 0.43 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr16_-_44512882 | 0.43 |
ENSDART00000191241
|
CR925804.2
|
|
| chr11_+_24339377 | 0.42 |
ENSDART00000133679
ENSDART00000135435 ENSDART00000017973 ENSDART00000131365 ENSDART00000186418 |
rbm39a
|
RNA binding motif protein 39a |
| chr12_-_35095414 | 0.40 |
ENSDART00000153229
|
si:dkey-21e13.3
|
si:dkey-21e13.3 |
| chr19_+_12237945 | 0.39 |
ENSDART00000190034
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr5_+_37903790 | 0.39 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr2_-_52020049 | 0.37 |
ENSDART00000128198
ENSDART00000135331 |
scn12aa
|
sodium channel, voltage gated, type XII, alpha a |
| chr24_-_21588914 | 0.36 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr2_-_50298337 | 0.35 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr23_+_3721042 | 0.35 |
ENSDART00000143323
|
smim29
|
small integral membrane protein 29 |
| chr7_+_63325819 | 0.32 |
ENSDART00000085612
ENSDART00000161436 |
pcdh7b
|
protocadherin 7b |
| chr13_+_25412307 | 0.31 |
ENSDART00000175339
|
calhm1
|
calcium homeostasis modulator 1 |
| chr15_+_40792667 | 0.31 |
ENSDART00000193186
ENSDART00000184559 |
fat3a
|
FAT atypical cadherin 3a |
| chr18_+_44080948 | 0.30 |
ENSDART00000191036
|
BX908388.2
|
|
| chr16_-_50157187 | 0.28 |
ENSDART00000155162
|
nek10
|
NIMA-related kinase 10 |
| chr24_-_18809433 | 0.28 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr4_-_5317006 | 0.27 |
ENSDART00000150867
|
si:ch211-214j24.15
|
si:ch211-214j24.15 |
| chr16_+_29630965 | 0.27 |
ENSDART00000185820
ENSDART00000192105 ENSDART00000153683 ENSDART00000186713 |
VPS72
|
si:ch211-203d17.1 |
| chr2_-_24270062 | 0.26 |
ENSDART00000192445
|
myh7
|
myosin heavy chain 7 |
| chr3_+_1724941 | 0.25 |
ENSDART00000193402
|
BX321875.2
|
|
| chr13_+_9678262 | 0.25 |
ENSDART00000111365
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr23_-_4019928 | 0.24 |
ENSDART00000021062
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr16_-_23797570 | 0.22 |
ENSDART00000077834
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr5_-_24112175 | 0.22 |
ENSDART00000131595
|
si:ch211-114c12.5
|
si:ch211-114c12.5 |
| chr6_+_52947186 | 0.22 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr8_+_7801060 | 0.21 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr5_+_13326765 | 0.21 |
ENSDART00000090851
|
ydjc
|
YdjC chitooligosaccharide deacetylase homolog |
| chr23_-_4019699 | 0.21 |
ENSDART00000159780
|
slc9a8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr19_-_15434813 | 0.20 |
ENSDART00000019843
|
ftr55
|
finTRIM family, member 55 |
| chr25_+_3788074 | 0.20 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr10_+_23441488 | 0.19 |
ENSDART00000100783
|
si:ch73-122g19.1
|
si:ch73-122g19.1 |
| chr3_-_34084387 | 0.17 |
ENSDART00000155365
|
ighv4-3
|
immunoglobulin heavy variable 4-3 |
| chr6_+_52947699 | 0.17 |
ENSDART00000180913
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_+_1602916 | 0.16 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr20_-_45058041 | 0.15 |
ENSDART00000124298
|
klhl29
|
kelch-like family member 29 |
| chr22_+_2778600 | 0.14 |
ENSDART00000134028
|
si:dkey-20i20.9
|
si:dkey-20i20.9 |
| chr20_-_44460789 | 0.12 |
ENSDART00000085442
|
mut
|
methylmalonyl CoA mutase |
| chr4_-_13502549 | 0.12 |
ENSDART00000140366
|
si:ch211-266a5.12
|
si:ch211-266a5.12 |
| chr15_+_3246475 | 0.12 |
ENSDART00000189565
|
LO017656.2
|
|
| chr4_+_74396786 | 0.12 |
ENSDART00000127501
ENSDART00000174347 |
zmp:0000001020
|
zmp:0000001020 |
| chr12_+_28854963 | 0.12 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr25_+_10947743 | 0.12 |
ENSDART00000073381
|
mhc1lfa
|
major histocompatibility complex class I LFA |
| chr15_-_38218320 | 0.11 |
ENSDART00000152363
|
rhoga
|
ras homolog family member Ga |
| chr17_-_6600899 | 0.10 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr17_+_4408809 | 0.10 |
ENSDART00000113401
|
si:zfos-364h11.1
|
si:zfos-364h11.1 |
| chr4_+_22613625 | 0.10 |
ENSDART00000131161
|
sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr5_-_26247973 | 0.10 |
ENSDART00000098527
|
erap1b
|
endoplasmic reticulum aminopeptidase 1b |
| chr7_-_36113098 | 0.09 |
ENSDART00000064913
|
fto
|
fat mass and obesity associated |
| chr4_+_73672430 | 0.08 |
ENSDART00000174310
ENSDART00000150505 |
si:dkey-262g12.7
|
si:dkey-262g12.7 |
| chr14_+_1355857 | 0.07 |
ENSDART00000188008
|
bbs12
|
Bardet-Biedl syndrome 12 |
| chr7_-_59515569 | 0.05 |
ENSDART00000163343
ENSDART00000165457 ENSDART00000163745 |
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr4_+_19127973 | 0.04 |
ENSDART00000136611
|
si:dkey-21o22.2
|
si:dkey-21o22.2 |
| chr12_+_19030391 | 0.03 |
ENSDART00000153927
|
si:ch73-139e5.2
|
si:ch73-139e5.2 |
| chr25_+_18953756 | 0.03 |
ENSDART00000154291
|
tdg.2
|
thymine DNA glycosylase, tandem duplicate 2 |
| chr1_+_11691722 | 0.03 |
ENSDART00000134531
|
eif3bb
|
eukaryotic translation initiation factor 3, subunit Bb |
| chr6_-_40352215 | 0.02 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr25_-_18953322 | 0.02 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr21_-_43094210 | 0.01 |
ENSDART00000144747
|
si:ch73-68b22.2
|
si:ch73-68b22.2 |
| chr7_+_2244859 | 0.01 |
ENSDART00000114242
ENSDART00000064292 |
si:ch211-285c6.4
|
si:ch211-285c6.4 |
| chr21_-_13972745 | 0.01 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr21_-_7940043 | 0.01 |
ENSDART00000099733
ENSDART00000136671 |
f2rl1.1
|
coagulation factor II (thrombin) receptor-like 1, tandem duplicate 1 |
| chr8_+_20440297 | 0.01 |
ENSDART00000189486
|
mknk2b
|
MAP kinase interacting serine/threonine kinase 2b |
| chr4_-_7811925 | 0.01 |
ENSDART00000103070
|
cdk17
|
cyclin-dependent kinase 17 |
| chr2_+_26240631 | 0.00 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of six3a+six3b+six7_six1a+six2a+six2b+six5+six9_six1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.4 | 1.3 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.3 | 1.4 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.3 | 3.5 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 8.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.2 | 2.8 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.2 | 1.9 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.2 | 2.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.7 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 0.1 | 0.4 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 1.6 | GO:0086001 | cardiac muscle cell action potential(GO:0086001) |
| 0.1 | 1.2 | GO:0033198 | response to ATP(GO:0033198) |
| 0.1 | 0.6 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.4 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.8 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.8 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 0.4 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 0.5 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.0 | 1.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.4 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.5 | GO:0071482 | cellular response to light stimulus(GO:0071482) |
| 0.0 | 0.9 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.6 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.1 | 1.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.4 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 2.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.8 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.6 | 1.9 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 1.8 | GO:0017020 | myosin phosphatase regulator activity(GO:0017020) |
| 0.3 | 1.4 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.3 | 3.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.3 | 0.8 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 1.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 8.9 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.7 | GO:0002058 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.1 | 0.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 1.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.1 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.4 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 2.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 2.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 1.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.1 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.4 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.1 | GO:0005126 | cytokine receptor binding(GO:0005126) |
| 0.0 | 0.8 | GO:0032934 | sterol binding(GO:0032934) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.3 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 4.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.5 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |