Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
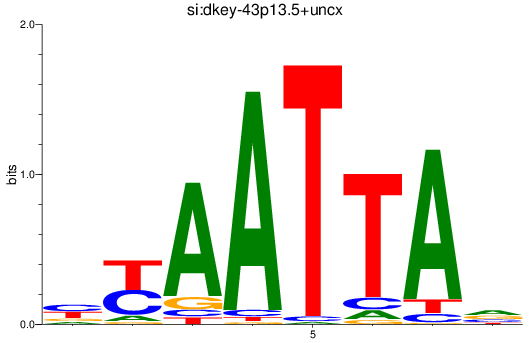
Results for si:dkey-43p13.5+uncx
Z-value: 1.19
Transcription factors associated with si:dkey-43p13.5+uncx
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
uncx
|
ENSDARG00000102976 | UNC homeobox |
|
si_dkey-43p13.5
|
ENSDARG00000104199 | si_dkey-43p13.5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-43p13.5 | dr11_v1_chr3_+_43086548_43086548 | 0.77 | 1.0e-04 | Click! |
| uncx | dr11_v1_chr3_-_43356082_43356082 | 0.18 | 4.7e-01 | Click! |
Activity profile of si:dkey-43p13.5+uncx motif
Sorted Z-values of si:dkey-43p13.5+uncx motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_55248496 | 4.10 |
ENSDART00000098615
|
nanos3
|
nanos homolog 3 |
| chr25_+_14507567 | 4.03 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr16_+_29509133 | 3.95 |
ENSDART00000112116
|
ctss2.1
|
cathepsin S, ortholog2, tandem duplicate 1 |
| chr19_+_15440841 | 3.47 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_-_35171768 | 3.24 |
ENSDART00000192896
|
traip
|
TRAF-interacting protein |
| chr24_+_38301080 | 3.22 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr16_+_54209504 | 3.07 |
ENSDART00000020033
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr17_-_16422654 | 3.05 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr19_+_15441022 | 2.99 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr8_+_30452945 | 2.91 |
ENSDART00000062303
|
foxd5
|
forkhead box D5 |
| chr18_+_619619 | 2.79 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr2_-_7246848 | 2.71 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr8_+_21353878 | 2.57 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr24_-_21923930 | 2.49 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr6_+_41191482 | 2.38 |
ENSDART00000000877
|
opn1mw3
|
opsin 1 (cone pigments), medium-wave-sensitive, 3 |
| chr18_-_33979693 | 2.33 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr3_-_60175470 | 2.32 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr23_-_18913032 | 2.28 |
ENSDART00000136678
|
si:ch211-209j10.6
|
si:ch211-209j10.6 |
| chr12_-_35830625 | 2.25 |
ENSDART00000180028
|
CU459056.1
|
|
| chr2_+_33326522 | 2.24 |
ENSDART00000056655
|
klf17
|
Kruppel-like factor 17 |
| chr21_-_37194365 | 2.23 |
ENSDART00000100286
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr11_-_35171162 | 2.22 |
ENSDART00000017393
|
traip
|
TRAF-interacting protein |
| chr19_+_12406583 | 2.20 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr23_-_31913069 | 2.13 |
ENSDART00000135526
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr12_-_43664682 | 2.12 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr4_-_52165969 | 2.10 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr3_-_23643751 | 2.10 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr17_-_14876758 | 2.08 |
ENSDART00000155857
|
nid2a
|
nidogen 2a (osteonidogen) |
| chr18_+_19456648 | 2.05 |
ENSDART00000079695
|
zwilch
|
zwilch kinetochore protein |
| chr2_+_20793982 | 2.04 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr21_+_15592426 | 2.03 |
ENSDART00000138207
|
smarcb1b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1b |
| chr23_-_31913231 | 2.03 |
ENSDART00000146852
ENSDART00000085054 |
mtfr2
|
mitochondrial fission regulator 2 |
| chr13_+_7442023 | 2.01 |
ENSDART00000080975
|
tnfaip2b
|
tumor necrosis factor, alpha-induced protein 2b |
| chr13_+_30903816 | 1.99 |
ENSDART00000191727
|
ercc6
|
excision repair cross-complementation group 6 |
| chr1_-_999556 | 1.96 |
ENSDART00000170884
ENSDART00000172235 |
gart
|
phosphoribosylglycinamide formyltransferase |
| chr8_+_23356264 | 1.94 |
ENSDART00000145062
|
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr22_-_15593824 | 1.93 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr8_-_50888806 | 1.92 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr1_-_45215343 | 1.92 |
ENSDART00000014727
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr23_-_17003533 | 1.92 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr3_-_4663602 | 1.88 |
ENSDART00000083532
|
slc25a38a
|
solute carrier family 25, member 38a |
| chr2_+_58841181 | 1.86 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr25_-_21031007 | 1.86 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr3_+_18807006 | 1.86 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr13_-_31296358 | 1.84 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr2_-_26596794 | 1.81 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr3_-_30488063 | 1.80 |
ENSDART00000055393
ENSDART00000151367 |
med25
|
mediator complex subunit 25 |
| chr14_+_22113331 | 1.80 |
ENSDART00000109759
|
tmx2a
|
thioredoxin-related transmembrane protein 2a |
| chr8_+_8936912 | 1.79 |
ENSDART00000135958
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr1_+_21725821 | 1.78 |
ENSDART00000132602
|
pax5
|
paired box 5 |
| chr1_-_33645967 | 1.77 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr2_-_17115256 | 1.77 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr17_+_30369396 | 1.75 |
ENSDART00000076611
|
greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr1_+_24557414 | 1.75 |
ENSDART00000076519
|
dctpp1
|
dCTP pyrophosphatase 1 |
| chr3_-_16719244 | 1.74 |
ENSDART00000055859
|
pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr16_-_7443388 | 1.74 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr7_+_56676838 | 1.72 |
ENSDART00000112242
|
zgc:194679
|
zgc:194679 |
| chr8_+_17168114 | 1.72 |
ENSDART00000183901
|
cenph
|
centromere protein H |
| chr22_+_18315490 | 1.70 |
ENSDART00000160413
|
gatad2ab
|
GATA zinc finger domain containing 2Ab |
| chr9_+_8396755 | 1.69 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr23_-_16485190 | 1.69 |
ENSDART00000155038
|
si:dkeyp-100a5.4
|
si:dkeyp-100a5.4 |
| chr15_-_14884332 | 1.69 |
ENSDART00000165237
|
si:ch211-24o8.4
|
si:ch211-24o8.4 |
| chr21_-_23331619 | 1.68 |
ENSDART00000007806
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr23_-_33775145 | 1.68 |
ENSDART00000132147
ENSDART00000027959 ENSDART00000160116 |
racgap1
|
Rac GTPase activating protein 1 |
| chr23_+_31913292 | 1.67 |
ENSDART00000136910
|
armc1l
|
armadillo repeat containing 1, like |
| chr10_+_16036246 | 1.65 |
ENSDART00000141586
ENSDART00000135868 ENSDART00000065037 ENSDART00000124502 |
lmnb1
|
lamin B1 |
| chr24_+_19415124 | 1.64 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr10_-_35257458 | 1.63 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr15_+_1534644 | 1.63 |
ENSDART00000130413
|
smc4
|
structural maintenance of chromosomes 4 |
| chr15_+_36309070 | 1.61 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr15_+_23799461 | 1.61 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr10_-_21362320 | 1.60 |
ENSDART00000189789
|
avd
|
avidin |
| chr8_+_23355484 | 1.60 |
ENSDART00000085361
ENSDART00000125729 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr5_+_66433287 | 1.60 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr5_-_20205075 | 1.59 |
ENSDART00000051611
|
dao.3
|
D-amino-acid oxidase, tandem duplicate 3 |
| chr17_-_48944465 | 1.59 |
ENSDART00000154110
|
si:ch1073-80i24.3
|
si:ch1073-80i24.3 |
| chr5_-_12219572 | 1.59 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr20_-_22476255 | 1.59 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr17_+_16046132 | 1.59 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr11_+_33818179 | 1.59 |
ENSDART00000109418
|
spoplb
|
speckle-type POZ protein-like b |
| chr20_-_27225876 | 1.58 |
ENSDART00000149204
ENSDART00000149732 |
si:dkey-85n7.7
|
si:dkey-85n7.7 |
| chr21_+_43404945 | 1.58 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr11_-_36341028 | 1.57 |
ENSDART00000146093
|
sort1a
|
sortilin 1a |
| chr3_+_45364849 | 1.57 |
ENSDART00000153974
|
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr1_+_513986 | 1.57 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr19_-_5669122 | 1.56 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr3_+_28860283 | 1.56 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr13_+_22295905 | 1.54 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr22_-_36530902 | 1.54 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr17_+_16046314 | 1.54 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_-_17693273 | 1.53 |
ENSDART00000146258
|
cfap97
|
cilia and flagella associated protein 97 |
| chr10_-_21362071 | 1.53 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_+_22485710 | 1.51 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr1_+_43686251 | 1.50 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr20_-_20930926 | 1.50 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr4_+_11723852 | 1.49 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr6_-_40778294 | 1.49 |
ENSDART00000019845
|
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr12_-_5728755 | 1.49 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr23_+_44374041 | 1.49 |
ENSDART00000136056
|
ephb4b
|
eph receptor B4b |
| chr14_-_33945692 | 1.48 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr22_+_16535575 | 1.48 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr14_-_38826739 | 1.48 |
ENSDART00000187633
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_30884706 | 1.48 |
ENSDART00000052126
|
yars
|
tyrosyl-tRNA synthetase |
| chr18_+_30998472 | 1.47 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr14_+_31496543 | 1.46 |
ENSDART00000170683
|
phf6
|
PHD finger protein 6 |
| chr16_+_14812585 | 1.44 |
ENSDART00000134087
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr13_+_27232848 | 1.44 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr19_-_5699703 | 1.43 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr13_-_31008275 | 1.42 |
ENSDART00000139394
|
wdfy4
|
WDFY family member 4 |
| chr12_+_16087077 | 1.42 |
ENSDART00000141898
|
znf281b
|
zinc finger protein 281b |
| chr19_+_29808471 | 1.41 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr16_+_42471455 | 1.41 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr14_-_22015232 | 1.40 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr7_+_24023653 | 1.40 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr8_+_28358161 | 1.40 |
ENSDART00000062682
|
adipor1b
|
adiponectin receptor 1b |
| chr5_+_26847190 | 1.39 |
ENSDART00000076742
|
imp4
|
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr23_+_11285662 | 1.38 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr2_+_15048410 | 1.38 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr4_+_28356606 | 1.37 |
ENSDART00000192995
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr9_-_32753535 | 1.37 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr11_-_41220794 | 1.36 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr20_-_2584101 | 1.36 |
ENSDART00000141595
ENSDART00000135760 |
med23
|
mediator complex subunit 23 |
| chr24_-_2450597 | 1.36 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr12_-_26415499 | 1.34 |
ENSDART00000185779
|
synpo2lb
|
synaptopodin 2-like b |
| chr17_-_49412313 | 1.34 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr20_+_4221978 | 1.34 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_+_56098590 | 1.33 |
ENSDART00000098453
|
cdh15
|
cadherin 15, type 1, M-cadherin (myotubule) |
| chr13_+_35637048 | 1.33 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr24_+_39638555 | 1.31 |
ENSDART00000078313
|
luc7l
|
LUC7-like (S. cerevisiae) |
| chr17_-_31695217 | 1.31 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr15_+_34988148 | 1.31 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr9_+_9927516 | 1.30 |
ENSDART00000161089
ENSDART00000168559 |
ttf2
|
transcription termination factor, RNA polymerase II |
| chr18_-_30499489 | 1.30 |
ENSDART00000033746
|
gins2
|
GINS complex subunit 2 |
| chr2_+_9970942 | 1.29 |
ENSDART00000153063
|
sec22ba
|
SEC22 homolog B, vesicle trafficking protein a |
| chr3_-_12930217 | 1.29 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr8_+_16758304 | 1.29 |
ENSDART00000133514
|
elovl7a
|
ELOVL fatty acid elongase 7a |
| chr20_+_25586099 | 1.29 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr21_+_723998 | 1.29 |
ENSDART00000160956
|
oaz1b
|
ornithine decarboxylase antizyme 1b |
| chr7_+_72003301 | 1.29 |
ENSDART00000012918
ENSDART00000182268 ENSDART00000185750 |
psmd9
|
proteasome 26S subunit, non-ATPase 9 |
| chr14_-_25935167 | 1.29 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr23_+_38251864 | 1.27 |
ENSDART00000183498
ENSDART00000129593 |
znf217
|
zinc finger protein 217 |
| chr13_-_12602920 | 1.27 |
ENSDART00000102311
|
lrit3b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 3b |
| chr11_+_17984354 | 1.26 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr23_+_39695827 | 1.26 |
ENSDART00000113893
ENSDART00000186679 |
tmco4
|
transmembrane and coiled-coil domains 4 |
| chr3_+_22442445 | 1.26 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr5_+_28497956 | 1.25 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr4_+_13901458 | 1.25 |
ENSDART00000137549
|
pphln1
|
periphilin 1 |
| chr17_+_8799451 | 1.25 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr2_+_41526904 | 1.25 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr6_-_8311044 | 1.25 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr3_+_45365098 | 1.25 |
ENSDART00000052746
ENSDART00000156555 |
ube2ia
|
ubiquitin-conjugating enzyme E2Ia |
| chr16_+_23961276 | 1.24 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr20_-_20931197 | 1.24 |
ENSDART00000152726
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr6_-_40922971 | 1.23 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr5_-_45958838 | 1.23 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr17_-_25649079 | 1.22 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr8_+_50953776 | 1.22 |
ENSDART00000013870
|
zgc:56596
|
zgc:56596 |
| chr5_+_60590796 | 1.21 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr23_+_1029450 | 1.21 |
ENSDART00000189196
|
si:zfos-905g2.1
|
si:zfos-905g2.1 |
| chr3_-_38918697 | 1.21 |
ENSDART00000145630
|
si:dkey-106c17.2
|
si:dkey-106c17.2 |
| chr10_-_22803740 | 1.21 |
ENSDART00000079469
ENSDART00000187968 ENSDART00000122543 |
pcolcea
|
procollagen C-endopeptidase enhancer a |
| chr19_+_6990970 | 1.21 |
ENSDART00000158758
ENSDART00000160482 ENSDART00000193566 |
kifc1
|
kinesin family member C1 |
| chr3_-_39695856 | 1.20 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr21_-_15200556 | 1.20 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr4_+_9177997 | 1.20 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr8_+_28259347 | 1.20 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr6_-_43283122 | 1.20 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr7_-_51368681 | 1.19 |
ENSDART00000146385
|
arhgap36
|
Rho GTPase activating protein 36 |
| chr5_-_41307550 | 1.19 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr20_+_13883131 | 1.19 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr17_-_11439815 | 1.18 |
ENSDART00000130105
|
psma3
|
proteasome subunit alpha 3 |
| chr3_+_13848226 | 1.18 |
ENSDART00000184342
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr17_+_24722646 | 1.18 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr2_+_6253246 | 1.17 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr10_+_7593185 | 1.17 |
ENSDART00000162617
ENSDART00000162590 ENSDART00000171744 |
ppp2cb
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr7_+_29167744 | 1.17 |
ENSDART00000076345
|
slc38a8b
|
solute carrier family 38, member 8b |
| chr15_-_3736773 | 1.17 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr3_+_53317040 | 1.16 |
ENSDART00000011780
|
xab2
|
XPA binding protein 2 |
| chr2_-_5723786 | 1.16 |
ENSDART00000100924
|
cwc25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr19_+_45962016 | 1.16 |
ENSDART00000169710
|
utp23
|
UTP23, small subunit (SSU) processome component, homolog (yeast) |
| chr21_+_43702016 | 1.16 |
ENSDART00000017176
|
dkc1
|
dyskeratosis congenita 1, dyskerin |
| chr10_+_16092671 | 1.15 |
ENSDART00000182761
ENSDART00000154835 |
megf10
|
multiple EGF-like-domains 10 |
| chr10_+_8197827 | 1.15 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr13_+_48359573 | 1.14 |
ENSDART00000161959
ENSDART00000165311 |
msh6
|
mutS homolog 6 (E. coli) |
| chr20_-_36408836 | 1.14 |
ENSDART00000076419
|
lbr
|
lamin B receptor |
| chr5_-_63302944 | 1.14 |
ENSDART00000047110
|
gsnb
|
gelsolin b |
| chr17_+_21295132 | 1.14 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr10_-_36691681 | 1.13 |
ENSDART00000122375
|
mrpl48
|
mitochondrial ribosomal protein L48 |
| chr23_-_24952264 | 1.13 |
ENSDART00000192085
|
zbtb48
|
zinc finger and BTB domain containing 48 |
| chr25_-_35956344 | 1.13 |
ENSDART00000066987
|
mphosph6
|
M-phase phosphoprotein 6 |
| chr6_-_8392104 | 1.13 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr19_-_7540821 | 1.13 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr20_-_37490612 | 1.12 |
ENSDART00000185458
|
si:ch211-202p1.5
|
si:ch211-202p1.5 |
| chr23_+_31912882 | 1.12 |
ENSDART00000140505
|
armc1l
|
armadillo repeat containing 1, like |
| chr7_+_22313533 | 1.11 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr10_+_17235370 | 1.11 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr11_-_30634286 | 1.11 |
ENSDART00000191019
|
zgc:153665
|
zgc:153665 |
| chr24_-_4450238 | 1.11 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr20_+_46513651 | 1.11 |
ENSDART00000152977
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr24_+_37800102 | 1.10 |
ENSDART00000187591
|
telo2
|
TEL2, telomere maintenance 2, homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-43p13.5+uncx
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 1.0 | 4.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.7 | 2.2 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.7 | 2.2 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.7 | 2.8 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.6 | 1.9 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.6 | 1.8 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.6 | 1.7 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.5 | 2.1 | GO:0048890 | epidermal cell fate specification(GO:0009957) lateral line ganglion development(GO:0048890) |
| 0.5 | 1.6 | GO:0098924 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.5 | 2.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 1.5 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.5 | 1.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 1.7 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 1.2 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.4 | 2.0 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.4 | 1.6 | GO:0009080 | alanine metabolic process(GO:0006522) alanine catabolic process(GO:0006524) pyruvate family amino acid metabolic process(GO:0009078) pyruvate family amino acid catabolic process(GO:0009080) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.4 | 2.0 | GO:0046084 | adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 0.4 | 1.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.4 | 1.9 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.4 | 1.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.4 | 2.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.4 | 1.5 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.4 | 3.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.4 | 1.1 | GO:0060898 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.4 | 1.1 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 2.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.4 | 1.1 | GO:0003097 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.4 | 1.1 | GO:1903334 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.3 | 3.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.3 | 1.0 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.3 | 1.6 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.3 | 1.6 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.3 | 1.9 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.3 | 1.3 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.3 | 1.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 0.9 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.3 | 0.9 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.3 | 0.8 | GO:0010658 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.3 | 1.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 1.6 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.3 | 1.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 1.0 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 0.8 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.3 | 1.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.3 | 1.3 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 0.7 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.2 | 1.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.2 | 2.2 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.2 | 3.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.2 | 1.0 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 2.1 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) |
| 0.2 | 0.9 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.2 | 0.9 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.2 | 4.7 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.2 | 1.1 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.2 | 0.7 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 0.7 | GO:0043416 | regulation of skeletal muscle tissue regeneration(GO:0043416) |
| 0.2 | 1.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.2 | 1.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 2.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.2 | 1.4 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.2 | 1.0 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.2 | 1.6 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.2 | 2.0 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.2 | 1.0 | GO:0060295 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.2 | 1.0 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.2 | 1.8 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.2 | 0.7 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.2 | 0.5 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.5 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.2 | 1.8 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.2 | 1.6 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.2 | 0.7 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 0.9 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 0.7 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 4.1 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.2 | 1.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.2 | 0.7 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.3 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.2 | 0.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 1.5 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 0.6 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 2.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 5.6 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.1 | 3.7 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 1.8 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.1 | 1.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.9 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.1 | 1.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.8 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 1.7 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.0 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.4 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 1.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 2.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.1 | 0.9 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 0.5 | GO:0089709 | L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 1.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.1 | 1.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.7 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.1 | 3.2 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 1.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.1 | 0.9 | GO:1903828 | negative regulation of cellular protein localization(GO:1903828) |
| 0.1 | 1.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.7 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 0.6 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.1 | 0.6 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.8 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.3 | GO:0031397 | negative regulation of protein ubiquitination(GO:0031397) |
| 0.1 | 0.4 | GO:0051958 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.1 | 0.8 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.1 | 1.3 | GO:1903318 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.1 | 1.7 | GO:0051383 | kinetochore assembly(GO:0051382) kinetochore organization(GO:0051383) |
| 0.1 | 0.9 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.9 | GO:0090224 | regulation of mitotic spindle organization(GO:0060236) regulation of spindle organization(GO:0090224) |
| 0.1 | 3.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 2.4 | GO:0021983 | pituitary gland development(GO:0021983) |
| 0.1 | 1.0 | GO:0044854 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.7 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.1 | 0.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 1.8 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.6 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 1.0 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.1 | 3.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.8 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 1.1 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 0.7 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 1.4 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.1 | GO:0097242 | regulation of nitric-oxide synthase activity(GO:0050999) positive regulation of nitric-oxide synthase activity(GO:0051000) beta-amyloid clearance(GO:0097242) regulation of beta-amyloid clearance(GO:1900221) |
| 0.1 | 1.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 1.1 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 2.3 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.1 | 0.9 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 1.7 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.9 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.6 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 0.2 | GO:1900180 | snRNA modification(GO:0040031) regulation of protein localization to nucleus(GO:1900180) |
| 0.1 | 0.9 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.9 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.4 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 1.9 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 2.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.9 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 0.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 1.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.2 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.7 | GO:1900052 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 0.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.2 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.1 | 3.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 1.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.1 | 1.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.4 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.1 | 0.7 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.1 | 0.6 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 1.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.5 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.5 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.8 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.1 | 1.8 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 0.6 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.3 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 1.0 | GO:0031935 | regulation of chromatin silencing(GO:0031935) |
| 0.1 | 0.2 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.1 | 1.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.1 | 1.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 2.1 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 1.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 | 0.3 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.8 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.5 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.3 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.9 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.9 | GO:0072599 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 1.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 1.5 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 2.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 1.1 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 2.9 | GO:0034504 | protein localization to nucleus(GO:0034504) |
| 0.0 | 0.3 | GO:0065005 | plasma lipoprotein particle assembly(GO:0034377) protein-lipid complex assembly(GO:0065005) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.6 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 2.5 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.0 | 0.2 | GO:0006154 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.5 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 1.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.3 | GO:0032917 | spermidine metabolic process(GO:0008216) polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 3.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 1.7 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 2.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.7 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 1.7 | GO:1902275 | regulation of chromatin organization(GO:1902275) |
| 0.0 | 0.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.8 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 3.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.8 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.4 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 1.3 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.6 | GO:2000404 | regulation of T cell migration(GO:2000404) |
| 0.0 | 0.0 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 1.1 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.4 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.1 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:2000780 | negative regulation of DNA repair(GO:0045738) negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 1.2 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.8 | GO:0006623 | protein targeting to vacuole(GO:0006623) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.8 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 2.9 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.6 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.0 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.0 | 0.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 1.4 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.5 | GO:0003401 | axis elongation(GO:0003401) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.4 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.7 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.3 | GO:0051654 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
| 0.0 | 0.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 8.4 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 2.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.5 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 1.1 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.8 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.8 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.7 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 1.1 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.1 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.4 | GO:0070672 | interleukin-15-mediated signaling pathway(GO:0035723) response to interleukin-15(GO:0070672) cellular response to interleukin-15(GO:0071350) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.0 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 1.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.4 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.5 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 1.0 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.6 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.8 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.3 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.0 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0002761 | regulation of myeloid leukocyte differentiation(GO:0002761) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0010324 | membrane invagination(GO:0010324) |
| 0.0 | 0.8 | GO:0031016 | pancreas development(GO:0031016) |
| 0.0 | 0.7 | GO:0002062 | chondrocyte differentiation(GO:0002062) |
| 0.0 | 5.7 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.0 | 0.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.9 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 1.4 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.3 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.3 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0007164 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 1.1 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.0 | GO:0018008 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.3 | GO:0097202 | activation of cysteine-type endopeptidase activity(GO:0097202) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.9 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.4 | 1.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.4 | 1.1 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.4 | 1.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.3 | 1.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.3 | 1.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.3 | 1.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.3 | 1.2 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.3 | 1.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.3 | 1.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.3 | 0.8 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.3 | 1.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 1.3 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 1.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.2 | 2.7 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 0.9 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 1.8 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 0.9 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.2 | 0.9 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.2 | 0.6 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.2 | 2.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.2 | 2.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.2 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 1.6 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.2 | 0.9 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.2 | 5.7 | GO:0043186 | P granule(GO:0043186) |
| 0.2 | 0.5 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.2 | 2.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.0 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.1 | 3.0 | GO:0090568 | nuclear transcriptional repressor complex(GO:0090568) |
| 0.1 | 1.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 1.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 1.0 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 2.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.9 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.3 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.1 | 1.0 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 1.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.4 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 5.1 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 0.5 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 2.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 2.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 1.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 0.8 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 0.3 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 0.5 | GO:0032021 | NELF complex(GO:0032021) |
| 0.1 | 0.4 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 1.6 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0043514 | interleukin-12 complex(GO:0043514) interleukin-23 complex(GO:0070743) |
| 0.0 | 2.1 | GO:0005761 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 2.5 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.6 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 4.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.7 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 1.6 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.8 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.6 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 2.4 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 1.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0033290 | eukaryotic 48S preinitiation complex(GO:0033290) |
| 0.0 | 2.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.3 | GO:0019867 | mitochondrial outer membrane(GO:0005741) outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 1.3 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 3.5 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 2.6 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.5 | GO:0036464 | cytoplasmic ribonucleoprotein granule(GO:0036464) |
| 0.0 | 0.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.5 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 2.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 4.7 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.1 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.0 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.2 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 0.4 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.0 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 4.1 | GO:0005694 | chromosome(GO:0005694) |
| 0.0 | 65.1 | GO:0005634 | nucleus(GO:0005634) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.9 | 2.6 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.7 | 2.8 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.6 | 3.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 5.8 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 2.6 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 1.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.4 | 1.6 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.4 | 1.6 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.4 | 1.1 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.4 | 1.1 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.4 | 3.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 1.8 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.4 | 1.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.3 | 2.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 1.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 1.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 0.9 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.3 | 1.7 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 0.9 | GO:0000035 | acyl binding(GO:0000035) |
| 0.3 | 0.9 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.3 | 0.8 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.3 | 1.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.3 | 1.0 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.2 | 1.7 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.2 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 1.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.2 | 0.7 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.2 | 0.7 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.2 | 2.0 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 0.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 4.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.2 | 1.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.2 | 1.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 1.0 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.2 | 1.3 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.2 | 0.6 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 1.7 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.5 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.2 | 1.9 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 2.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 1.7 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.2 | 0.8 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.2 | 1.6 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 0.6 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 2.6 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.8 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.4 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 2.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 1.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 1.9 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.1 | 1.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.1 | 1.7 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 2.6 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.6 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 5.2 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 0.4 | GO:0008518 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.1 | 0.3 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.1 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 1.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.4 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.5 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 1.4 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.6 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 1.0 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.3 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.1 | 1.8 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 0.8 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 2.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 1.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 1.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 3.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 6.5 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.8 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 1.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.9 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.3 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 0.6 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.6 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.1 | 2.8 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.1 | 0.3 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 1.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.5 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 3.3 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 0.7 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.1 | 1.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.1 | 1.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.7 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 1.6 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 1.1 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 2.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 1.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 1.4 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.3 | GO:0046966 | vitamin D receptor binding(GO:0042809) thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 0.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.4 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.0 | 1.0 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.2 | GO:0042164 | interleukin-12 binding(GO:0019972) interleukin-12 alpha subunit binding(GO:0042164) |
| 0.0 | 2.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.5 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.3 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.7 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.6 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 3.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.2 | GO:0043394 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 2.0 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.0 | 0.3 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.7 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.6 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 2.0 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.7 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0016793 | dGTPase activity(GO:0008832) triphosphoric monoester hydrolase activity(GO:0016793) |
| 0.0 | 0.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 2.6 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.9 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.6 | GO:0051119 | sugar transmembrane transporter activity(GO:0051119) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 2.0 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.4 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.0 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 3.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.5 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 35.6 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 2.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 1.7 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.8 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.0 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.5 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 5.1 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 8.2 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.9 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 0.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 1.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.3 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 1.0 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.1 | 1.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 0.8 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.9 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.8 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.9 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.1 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.0 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 1.1 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.7 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 3.4 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.1 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 3.1 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.1 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.2 | 1.7 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.2 | 5.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 4.1 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 3.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 9.0 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 1.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 3.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 5.3 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 1.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.1 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 1.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 2.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.6 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 0.8 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.9 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.1 | 1.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.1 | 0.7 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 0.9 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.1 | 1.4 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 0.8 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.1 | 1.1 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.2 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.6 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.5 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 1.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.6 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 1.2 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.9 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.3 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.8 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.7 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.1 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |