Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
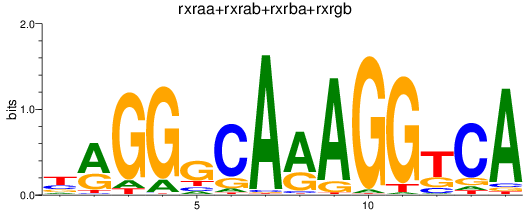
Results for rxraa+rxrab+rxrba+rxrgb
Z-value: 1.39
Transcription factors associated with rxraa+rxrab+rxrba+rxrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rxrgb
|
ENSDARG00000004697 | retinoid X receptor, gamma b |
|
rxrab
|
ENSDARG00000035127 | retinoid x receptor, alpha b |
|
rxraa
|
ENSDARG00000057737 | retinoid X receptor, alpha a |
|
rxrba
|
ENSDARG00000078954 | retinoid x receptor, beta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rxrba | dr11_v1_chr19_-_7274844_7274844 | 0.92 | 1.7e-08 | Click! |
| rxraa | dr11_v1_chr21_+_17880511_17880511 | 0.86 | 2.8e-06 | Click! |
| rxrab | dr11_v1_chr5_-_64511428_64511428 | 0.66 | 2.2e-03 | Click! |
| rxrgb | dr11_v1_chr20_+_33875256_33875256 | -0.05 | 8.3e-01 | Click! |
Activity profile of rxraa+rxrab+rxrba+rxrgb motif
Sorted Z-values of rxraa+rxrab+rxrba+rxrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_29996344 | 4.58 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr17_+_450956 | 4.53 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr10_+_45128375 | 4.03 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr24_-_29586082 | 3.58 |
ENSDART00000136763
|
vav3a
|
vav 3 guanine nucleotide exchange factor a |
| chr20_-_25522911 | 3.52 |
ENSDART00000063058
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr23_-_45504991 | 3.49 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr19_-_20093341 | 3.29 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr8_-_37263524 | 3.26 |
ENSDART00000061327
|
rh50
|
Rh50-like protein |
| chr15_+_47386939 | 3.19 |
ENSDART00000128224
|
FO904873.1
|
|
| chr2_-_57076687 | 3.16 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr5_-_36549024 | 3.08 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr16_-_2558653 | 3.05 |
ENSDART00000110365
|
adcy3a
|
adenylate cyclase 3a |
| chr18_-_35736591 | 2.77 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr3_+_3810919 | 2.74 |
ENSDART00000056035
|
FQ311927.1
|
|
| chr25_-_19420949 | 2.70 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr14_-_46238186 | 2.54 |
ENSDART00000173245
|
si:ch211-113d11.6
|
si:ch211-113d11.6 |
| chr21_-_1972236 | 2.46 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr23_-_1587955 | 2.29 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr20_-_1635922 | 2.19 |
ENSDART00000181502
|
CR846082.1
|
|
| chr1_-_9641845 | 2.17 |
ENSDART00000121490
ENSDART00000159411 |
ugt5b2
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B2 UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr1_+_25848231 | 2.16 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr24_+_39137001 | 2.16 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr19_+_41701660 | 2.13 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr17_+_30450163 | 2.07 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr5_-_69940868 | 2.03 |
ENSDART00000185924
ENSDART00000097357 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr23_+_27912079 | 2.00 |
ENSDART00000171859
|
BX539336.1
|
|
| chr17_+_12408405 | 2.00 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr25_-_30429607 | 1.97 |
ENSDART00000162429
ENSDART00000176535 |
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr8_-_7847921 | 1.97 |
ENSDART00000188094
|
zgc:113363
|
zgc:113363 |
| chr6_-_49547680 | 1.90 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr25_+_36347126 | 1.89 |
ENSDART00000152449
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr19_+_7043634 | 1.88 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr7_-_24520866 | 1.88 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr11_-_22303678 | 1.86 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr4_+_1283068 | 1.86 |
ENSDART00000167233
|
chrm2a
|
cholinergic receptor, muscarinic 2a |
| chr12_+_31673588 | 1.85 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr3_+_12732382 | 1.77 |
ENSDART00000158403
|
cyp2k19
|
cytochrome P450, family 2, subfamily k, polypeptide 19 |
| chr12_-_31457301 | 1.75 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr3_-_18805225 | 1.73 |
ENSDART00000133471
ENSDART00000131758 |
msrb1a
|
methionine sulfoxide reductase B1a |
| chr20_+_13175379 | 1.70 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr11_-_36957127 | 1.70 |
ENSDART00000168528
|
cacna1da
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, a |
| chr22_+_38778649 | 1.67 |
ENSDART00000075873
|
alpi.2
|
alkaline phosphatase, intestinal, tandem duplicate 2 |
| chr4_-_8152746 | 1.63 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr3_+_30257582 | 1.60 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr10_+_20180163 | 1.58 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr18_+_44769027 | 1.57 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr23_-_637347 | 1.56 |
ENSDART00000132175
|
l1camb
|
L1 cell adhesion molecule, paralog b |
| chr2_-_8102169 | 1.56 |
ENSDART00000131955
|
pls1
|
plastin 1 (I isoform) |
| chr23_+_2778813 | 1.55 |
ENSDART00000142621
|
top1
|
DNA topoisomerase I |
| chr4_-_31064105 | 1.54 |
ENSDART00000157670
|
si:dkey-11n14.1
|
si:dkey-11n14.1 |
| chr5_+_8919698 | 1.54 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr13_+_50375800 | 1.52 |
ENSDART00000099537
|
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr15_+_47618221 | 1.51 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr3_-_5016893 | 1.48 |
ENSDART00000165968
|
CSDC2 (1 of many)
|
cold shock domain containing C2 |
| chr6_+_43353271 | 1.48 |
ENSDART00000191775
|
BX927362.2
|
|
| chr7_+_73649686 | 1.48 |
ENSDART00000185589
|
PCP4L1
|
si:dkey-46i9.1 |
| chr3_+_12744083 | 1.48 |
ENSDART00000158554
ENSDART00000169545 |
cyp2k21
|
cytochrome P450, family 2, subfamily k, polypeptide 21 |
| chr8_+_999421 | 1.47 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr23_-_46126444 | 1.46 |
ENSDART00000030004
|
CABZ01071903.1
|
|
| chr11_+_36989696 | 1.45 |
ENSDART00000045888
|
tkta
|
transketolase a |
| chr23_+_42434348 | 1.44 |
ENSDART00000161027
|
cyp2aa1
|
cytochrome P450, family 2, subfamily AA, polypeptide 1 |
| chr1_-_46859398 | 1.44 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr7_+_34297271 | 1.43 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr3_+_4346854 | 1.43 |
ENSDART00000004273
|
si:dkey-73p2.3
|
si:dkey-73p2.3 |
| chr1_+_58094551 | 1.43 |
ENSDART00000146316
|
si:ch211-114l13.1
|
si:ch211-114l13.1 |
| chr20_+_43379029 | 1.43 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr6_+_60055168 | 1.42 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr20_+_1088658 | 1.41 |
ENSDART00000162991
|
BX537249.1
|
|
| chr24_-_1341543 | 1.40 |
ENSDART00000169341
|
nrp1a
|
neuropilin 1a |
| chr4_+_15944245 | 1.38 |
ENSDART00000134594
|
si:dkey-117n7.3
|
si:dkey-117n7.3 |
| chr12_-_34713888 | 1.38 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr15_+_45640906 | 1.36 |
ENSDART00000149361
ENSDART00000149079 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr25_-_19374710 | 1.35 |
ENSDART00000184483
ENSDART00000188706 |
map1ab
|
microtubule-associated protein 1Ab |
| chr14_-_2203124 | 1.35 |
ENSDART00000124171
|
si:dkeyp-115d7.2
|
si:dkeyp-115d7.2 |
| chr19_-_22498629 | 1.32 |
ENSDART00000185370
|
pleca
|
plectin a |
| chr4_+_2655358 | 1.30 |
ENSDART00000007638
|
bcap29
|
B cell receptor associated protein 29 |
| chr6_-_56877366 | 1.28 |
ENSDART00000168821
|
CDH22
|
cadherin 22 |
| chr21_+_1119046 | 1.28 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr18_-_12957451 | 1.26 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr7_+_34296789 | 1.26 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr12_+_1286642 | 1.26 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr9_-_37613792 | 1.25 |
ENSDART00000138345
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr7_-_50604626 | 1.24 |
ENSDART00000073903
ENSDART00000174031 |
crtc3
|
CREB regulated transcription coactivator 3 |
| chr13_-_44630111 | 1.24 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr7_+_65398161 | 1.23 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr21_+_10076203 | 1.22 |
ENSDART00000190383
|
CABZ01043506.1
|
|
| chr2_+_57801960 | 1.21 |
ENSDART00000147966
|
si:dkeyp-68b7.10
|
si:dkeyp-68b7.10 |
| chr19_-_41213718 | 1.18 |
ENSDART00000077121
|
pdk4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr8_+_19624589 | 1.16 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr17_+_12408188 | 1.16 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr11_+_24957858 | 1.16 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr24_-_11446156 | 1.14 |
ENSDART00000143921
ENSDART00000066778 |
acad11
|
acyl-CoA dehydrogenase family, member 11 |
| chr23_+_19790962 | 1.14 |
ENSDART00000142228
|
flna
|
filamin A, alpha (actin binding protein 280) |
| chr2_-_32688905 | 1.13 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr18_+_27926839 | 1.11 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr9_-_22834860 | 1.09 |
ENSDART00000146486
|
neb
|
nebulin |
| chr6_-_48082525 | 1.09 |
ENSDART00000192049
|
slc2a1b
|
solute carrier family 2 (facilitated glucose transporter), member 1b |
| chr20_-_24443680 | 1.09 |
ENSDART00000191337
|
CR385053.3
|
|
| chr1_-_17797802 | 1.08 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr23_+_44633858 | 1.07 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr1_+_20069535 | 1.06 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr18_+_44768829 | 1.06 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr20_-_42241150 | 1.04 |
ENSDART00000142791
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr15_-_1843831 | 1.04 |
ENSDART00000156718
ENSDART00000154175 |
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr6_-_59563597 | 1.04 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr22_-_17953529 | 1.03 |
ENSDART00000135716
|
ncanb
|
neurocan b |
| chr2_+_52847049 | 1.03 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr3_+_59117136 | 1.03 |
ENSDART00000182745
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr3_+_1749793 | 1.03 |
ENSDART00000149308
|
si:dkeyp-52c3.7
|
si:dkeyp-52c3.7 |
| chr11_-_45135643 | 1.01 |
ENSDART00000170863
|
cant1b
|
calcium activated nucleotidase 1b |
| chr8_+_8012570 | 1.00 |
ENSDART00000183429
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr7_-_52096498 | 0.98 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr25_-_35113891 | 0.98 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr20_-_1239596 | 0.97 |
ENSDART00000049063
ENSDART00000140650 |
ankrd6b
|
ankyrin repeat domain 6b |
| chr17_-_37184655 | 0.96 |
ENSDART00000180447
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr25_-_21085661 | 0.96 |
ENSDART00000099355
|
prr5a
|
proline rich 5a (renal) |
| chr13_-_37254777 | 0.95 |
ENSDART00000139734
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr7_+_20629411 | 0.95 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr12_+_49125510 | 0.94 |
ENSDART00000185804
|
FO704607.1
|
|
| chr18_+_9635178 | 0.94 |
ENSDART00000183486
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr12_+_26670778 | 0.93 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr10_-_17284055 | 0.93 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr11_-_6974022 | 0.92 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr8_-_43716897 | 0.91 |
ENSDART00000163237
|
ep400
|
E1A binding protein p400 |
| chr2_-_26476030 | 0.90 |
ENSDART00000145262
ENSDART00000132125 |
acadm
|
acyl-CoA dehydrogenase medium chain |
| chr4_-_858434 | 0.90 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr1_+_38858399 | 0.90 |
ENSDART00000165454
|
CU915762.1
|
|
| chr13_+_844150 | 0.90 |
ENSDART00000058260
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr9_-_7212973 | 0.90 |
ENSDART00000133638
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr13_-_34683370 | 0.90 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr8_+_32742650 | 0.90 |
ENSDART00000138117
|
hmcn2
|
hemicentin 2 |
| chr22_+_15323930 | 0.88 |
ENSDART00000142416
|
si:dkey-236e20.3
|
si:dkey-236e20.3 |
| chr16_-_22225295 | 0.87 |
ENSDART00000163519
|
LO017682.1
|
|
| chr16_+_10329701 | 0.87 |
ENSDART00000172845
|
mdc1
|
mediator of DNA damage checkpoint 1 |
| chr5_-_68927728 | 0.87 |
ENSDART00000132838
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr17_-_608857 | 0.87 |
ENSDART00000163431
|
KLHL28
|
kelch like family member 28 |
| chr8_-_12468744 | 0.85 |
ENSDART00000135019
|
FIBCD1 (1 of many)
|
si:dkeyp-51b7.3 |
| chr15_-_16946124 | 0.85 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr18_-_2727764 | 0.83 |
ENSDART00000160841
|
ARHGEF17
|
si:ch211-248g20.5 |
| chr1_-_1627487 | 0.83 |
ENSDART00000166094
|
clic6
|
chloride intracellular channel 6 |
| chr3_+_1179601 | 0.83 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr13_-_41546779 | 0.83 |
ENSDART00000163331
|
pcdh15a
|
protocadherin-related 15a |
| chr13_+_43050562 | 0.83 |
ENSDART00000016602
|
cdh23
|
cadherin-related 23 |
| chr23_+_2760573 | 0.82 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr22_-_3595439 | 0.82 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr18_+_27515640 | 0.80 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr3_-_55531450 | 0.80 |
ENSDART00000155376
|
tex2
|
testis expressed 2 |
| chr20_+_13969414 | 0.80 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr17_-_8169774 | 0.80 |
ENSDART00000091828
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr5_-_37933983 | 0.79 |
ENSDART00000185364
|
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr24_-_32522587 | 0.79 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr13_-_31878043 | 0.79 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr23_-_12788113 | 0.79 |
ENSDART00000103231
ENSDART00000146640 ENSDART00000180549 |
si:dkey-96f10.1
|
si:dkey-96f10.1 |
| chr8_-_43721359 | 0.78 |
ENSDART00000166581
ENSDART00000171633 |
ep400
|
E1A binding protein p400 |
| chr18_+_25546227 | 0.78 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr14_+_6429399 | 0.78 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr12_-_36260532 | 0.78 |
ENSDART00000022533
|
kcnj2a
|
potassium inwardly-rectifying channel, subfamily J, member 2a |
| chr7_+_24520518 | 0.77 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr17_-_15777350 | 0.77 |
ENSDART00000155711
|
ankrd6a
|
ankyrin repeat domain 6a |
| chr12_-_1694053 | 0.76 |
ENSDART00000169370
|
CABZ01081830.2
|
|
| chr7_-_51953807 | 0.76 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr12_-_36268723 | 0.76 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr8_-_46700278 | 0.75 |
ENSDART00000143780
|
gpr153
|
G protein-coupled receptor 153 |
| chr18_-_15771551 | 0.75 |
ENSDART00000130931
ENSDART00000154079 |
si:ch211-219a15.3
|
si:ch211-219a15.3 |
| chr25_+_22017182 | 0.75 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr24_-_32150276 | 0.75 |
ENSDART00000166212
|
cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr6_-_21830405 | 0.74 |
ENSDART00000151803
ENSDART00000113497 |
setd5
|
SET domain containing 5 |
| chr19_+_3056450 | 0.74 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr15_+_28268135 | 0.73 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr11_-_35970966 | 0.73 |
ENSDART00000188805
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr21_+_15883546 | 0.73 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr20_-_7128612 | 0.72 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr9_-_374693 | 0.72 |
ENSDART00000166571
|
si:dkey-11f4.7
|
si:dkey-11f4.7 |
| chr3_+_12718100 | 0.72 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr8_-_16464453 | 0.72 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr25_-_204019 | 0.71 |
ENSDART00000188440
ENSDART00000191735 |
FP236318.2
|
|
| chr12_-_6033824 | 0.70 |
ENSDART00000131301
ENSDART00000139419 ENSDART00000032050 |
g6pca.1
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 1 |
| chr1_-_44314 | 0.70 |
ENSDART00000160050
|
tmem39a
|
transmembrane protein 39A |
| chr6_+_154556 | 0.69 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr18_+_14342326 | 0.68 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr13_-_31878263 | 0.68 |
ENSDART00000180411
|
syt14a
|
synaptotagmin XIVa |
| chr23_-_44903048 | 0.68 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr8_+_51084993 | 0.68 |
ENSDART00000127709
ENSDART00000053768 |
DISP3
|
si:dkey-32e23.6 |
| chr6_+_20651398 | 0.68 |
ENSDART00000183663
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr14_-_34700633 | 0.67 |
ENSDART00000150358
|
ablim3
|
actin binding LIM protein family, member 3 |
| chr12_-_46112892 | 0.67 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr3_+_7808459 | 0.67 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr9_+_41224100 | 0.67 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr5_-_23362602 | 0.67 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr1_-_50167792 | 0.66 |
ENSDART00000182993
|
BX537118.1
|
|
| chr3_-_55197881 | 0.66 |
ENSDART00000179903
|
kank2
|
KN motif and ankyrin repeat domains 2 |
| chr15_+_19797918 | 0.65 |
ENSDART00000113314
|
si:ch211-229d2.5
|
si:ch211-229d2.5 |
| chr2_-_32501501 | 0.64 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr11_-_31234584 | 0.64 |
ENSDART00000170700
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr18_+_12655766 | 0.64 |
ENSDART00000144246
|
tbxas1
|
thromboxane A synthase 1 (platelet) |
| chr2_-_39017838 | 0.63 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr21_-_5879897 | 0.63 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr16_-_19568388 | 0.62 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr19_+_14921000 | 0.62 |
ENSDART00000144052
|
oprd1a
|
opioid receptor, delta 1a |
| chr23_-_46040618 | 0.62 |
ENSDART00000161415
|
CABZ01080918.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of rxraa+rxrab+rxrba+rxrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0009750 | response to fructose(GO:0009750) |
| 0.7 | 2.6 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.6 | 1.7 | GO:0015911 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.6 | 3.5 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.6 | 2.8 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.5 | 1.9 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.4 | 3.3 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.4 | 1.2 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.4 | 1.4 | GO:0019626 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.3 | 1.4 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 1.4 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.3 | 1.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 3.6 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.2 | 0.7 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.2 | 0.7 | GO:0071305 | cellular response to vitamin D(GO:0071305) |
| 0.2 | 0.7 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.2 | 1.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 9.6 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.2 | 1.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.2 | 0.5 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.2 | 1.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.2 | 1.5 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 4.3 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.2 | 1.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.2 | 0.6 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 0.9 | GO:0051793 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.1 | 2.4 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.0 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.5 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 0.8 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.9 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 1.3 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 1.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 1.8 | GO:1902307 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 4.0 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.8 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.5 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.9 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 1.9 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 1.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 1.7 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.7 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 0.8 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 1.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 2.1 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.1 | 2.1 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 1.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 1.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.1 | 0.3 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.3 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.8 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.1 | 0.8 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.1 | 2.8 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.1 | 3.0 | GO:0006171 | cAMP biosynthetic process(GO:0006171) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.7 | GO:0071357 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0039531 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.4 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 0.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 0.1 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.0 | 0.5 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.8 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 1.4 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 3.9 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 1.9 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.7 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 2.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.2 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.1 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0019883 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 1.0 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.7 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 1.0 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 1.0 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.8 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0099590 | postsynaptic neurotransmitter receptor internalization(GO:0098884) neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.7 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.2 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.3 | GO:0046058 | cAMP metabolic process(GO:0046058) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.6 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 1.1 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.5 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.9 | 2.6 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.4 | 2.8 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.4 | 1.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 1.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.2 | 1.4 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.2 | 0.8 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 0.8 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.6 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.4 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.1 | 2.1 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.8 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.9 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 1.9 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 1.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 3.6 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.8 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 2.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.2 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.1 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 2.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 2.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 1.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 1.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.2 | GO:0031968 | outer membrane(GO:0019867) organelle outer membrane(GO:0031968) |
| 0.0 | 0.4 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.9 | 2.7 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.9 | 2.6 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.6 | 2.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.5 | 1.4 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.4 | 1.7 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.3 | 1.2 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.2 | 3.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.4 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.2 | 0.7 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.2 | 3.0 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.2 | 1.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.2 | 0.6 | GO:0038046 | enkephalin receptor activity(GO:0038046) |
| 0.2 | 2.4 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 1.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.2 | 11.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.2 | 0.5 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 1.7 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.2 | 1.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 0.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.1 | 1.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 3.3 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 1.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.1 | 1.8 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 3.0 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.1 | 4.1 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 1.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.9 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 1.7 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.6 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.1 | 0.3 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 4.0 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.5 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.8 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 4.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.8 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.3 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 1.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 1.5 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.9 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 5.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.0 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.3 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.0 | 1.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.4 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 1.9 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 1.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 1.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 3.8 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.1 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 2.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 3.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 1.0 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 0.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 3.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.9 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.7 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 2.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |