Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
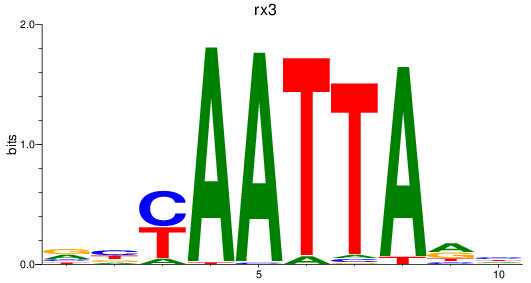
Results for rx3
Z-value: 0.72
Transcription factors associated with rx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx3
|
ENSDARG00000052893 | retinal homeobox gene 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx3 | dr11_v1_chr21_+_10756154_10756154 | 0.14 | 5.6e-01 | Click! |
Activity profile of rx3 motif
Sorted Z-values of rx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_10503043 | 2.24 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr11_+_30244356 | 2.23 |
ENSDART00000036050
ENSDART00000150080 |
rs1a
|
retinoschisin 1a |
| chr19_+_10339538 | 1.82 |
ENSDART00000151808
ENSDART00000151235 |
rcvrn3
|
recoverin 3 |
| chr5_-_50992690 | 1.75 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr20_-_40755614 | 1.58 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr3_-_50443607 | 1.47 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr10_-_7756865 | 1.46 |
ENSDART00000114373
ENSDART00000125407 ENSDART00000016317 |
loxa
|
lysyl oxidase a |
| chr2_+_39021282 | 1.41 |
ENSDART00000056577
|
RBP1 (1 of many)
|
si:ch211-119o8.7 |
| chr13_+_28701233 | 1.33 |
ENSDART00000135931
|
si:ch211-67n3.9
|
si:ch211-67n3.9 |
| chr14_-_2933185 | 1.29 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr21_-_42100471 | 1.20 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr22_+_28337204 | 1.08 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr22_+_28337429 | 0.92 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr23_-_19230627 | 0.91 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr13_-_44630111 | 0.88 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr14_+_40874608 | 0.88 |
ENSDART00000168448
|
si:ch211-106m9.1
|
si:ch211-106m9.1 |
| chr10_-_13343831 | 0.84 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr11_+_38280454 | 0.83 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr2_+_22702488 | 0.83 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr5_+_64732036 | 0.73 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr18_-_43884044 | 0.71 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr17_-_200316 | 0.69 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr9_-_21912227 | 0.66 |
ENSDART00000145576
|
lmo7a
|
LIM domain 7a |
| chr4_-_4834347 | 0.64 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr1_-_50859053 | 0.64 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr3_-_61162750 | 0.63 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr4_-_4834617 | 0.62 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr20_+_34717403 | 0.62 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr18_-_5598958 | 0.61 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr4_+_21129752 | 0.56 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr15_-_21877726 | 0.55 |
ENSDART00000127819
ENSDART00000145646 ENSDART00000100897 ENSDART00000144739 |
zgc:162608
|
zgc:162608 |
| chr23_-_1348933 | 0.54 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr21_-_22827548 | 0.53 |
ENSDART00000079161
|
angptl5
|
angiopoietin-like 5 |
| chr10_+_42423318 | 0.52 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr5_+_64732270 | 0.47 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr11_-_44801968 | 0.47 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr12_+_24342303 | 0.46 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr12_-_6880694 | 0.45 |
ENSDART00000171846
|
pcdh15b
|
protocadherin-related 15b |
| chr8_+_7801060 | 0.43 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr9_+_50001746 | 0.42 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr22_-_7129631 | 0.41 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr6_+_24398907 | 0.41 |
ENSDART00000167482
|
tgfbr3
|
transforming growth factor, beta receptor III |
| chr24_-_39518599 | 0.41 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr22_+_5176693 | 0.41 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr8_+_20776654 | 0.40 |
ENSDART00000135850
|
nfic
|
nuclear factor I/C |
| chr10_+_21867307 | 0.37 |
ENSDART00000126629
|
cbln17
|
cerebellin 17 |
| chr4_-_20292821 | 0.36 |
ENSDART00000136069
ENSDART00000192504 |
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr7_-_23768234 | 0.36 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr24_+_13316737 | 0.36 |
ENSDART00000191658
|
SBSPON
|
somatomedin B and thrombospondin type 1 domain containing |
| chr16_+_46111849 | 0.35 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr25_-_27621268 | 0.35 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr3_-_23406964 | 0.35 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr22_-_21897203 | 0.34 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr11_-_42554290 | 0.34 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr5_-_67629263 | 0.33 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr22_+_5176255 | 0.32 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr17_-_40956035 | 0.32 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr15_-_22074315 | 0.31 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr10_-_32494304 | 0.30 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr20_+_28861629 | 0.30 |
ENSDART00000187274
ENSDART00000047826 |
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr1_-_513762 | 0.29 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr4_+_3980247 | 0.29 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr14_-_858985 | 0.28 |
ENSDART00000148687
ENSDART00000149375 |
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr13_+_38814521 | 0.28 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr14_+_30795559 | 0.28 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr24_-_31425799 | 0.27 |
ENSDART00000157998
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr18_-_2433011 | 0.26 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr6_-_6487876 | 0.26 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr16_+_33902006 | 0.24 |
ENSDART00000161807
ENSDART00000159474 |
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr8_-_24252933 | 0.24 |
ENSDART00000057624
|
zgc:110353
|
zgc:110353 |
| chr11_-_30158191 | 0.24 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr1_+_18811679 | 0.23 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr20_+_28861435 | 0.23 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr22_+_18477934 | 0.22 |
ENSDART00000132684
|
cilp2
|
cartilage intermediate layer protein 2 |
| chr25_-_13842618 | 0.22 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr17_+_19630272 | 0.21 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr15_-_14552101 | 0.21 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr19_-_30524952 | 0.20 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr1_-_18811517 | 0.20 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr9_+_22485343 | 0.20 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr10_-_32494499 | 0.19 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr8_+_6410933 | 0.19 |
ENSDART00000168789
|
lrrtm4l2
|
leucine rich repeat transmembrane neuronal 4 like 2 |
| chr9_-_50001606 | 0.18 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr12_+_48803098 | 0.17 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr2_+_40294313 | 0.17 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr7_+_34592526 | 0.17 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr8_+_25034544 | 0.16 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr21_-_35419486 | 0.16 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr14_+_36521005 | 0.15 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr24_-_35282568 | 0.14 |
ENSDART00000167406
ENSDART00000088609 |
sntg1
|
syntrophin, gamma 1 |
| chr13_-_31017960 | 0.14 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr1_-_17715493 | 0.13 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr21_+_32820175 | 0.13 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr24_-_4782052 | 0.13 |
ENSDART00000149911
|
agtr1b
|
angiotensin II receptor, type 1b |
| chr11_-_29768054 | 0.13 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr18_+_2228737 | 0.12 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr22_-_10121880 | 0.12 |
ENSDART00000002348
|
rdh5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr22_-_8725768 | 0.11 |
ENSDART00000189873
ENSDART00000181819 |
si:ch73-27e22.1
si:ch73-27e22.8
|
si:ch73-27e22.1 si:ch73-27e22.8 |
| chr20_+_28803977 | 0.11 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr15_+_5360407 | 0.11 |
ENSDART00000110420
|
or112-1
|
odorant receptor, family A, subfamily 112, member 1 |
| chr14_-_7207961 | 0.10 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr5_-_27438812 | 0.10 |
ENSDART00000078755
|
drd7
|
dopamine receptor D7 |
| chr14_+_23717165 | 0.09 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr19_-_42391383 | 0.08 |
ENSDART00000110075
ENSDART00000087002 |
plekho1a
|
pleckstrin homology domain containing, family O member 1a |
| chr4_-_19982119 | 0.08 |
ENSDART00000169248
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr13_+_38430466 | 0.08 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr6_-_40352215 | 0.07 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr12_+_31744217 | 0.07 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr10_-_33297864 | 0.06 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr14_-_8940499 | 0.05 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr21_+_25777425 | 0.03 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr21_-_32060993 | 0.03 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr19_-_8768564 | 0.03 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr2_-_15324837 | 0.02 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr14_+_8940326 | 0.02 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr9_+_22364997 | 0.01 |
ENSDART00000188054
ENSDART00000046116 |
crygs3
|
crystallin, gamma S3 |
| chr11_+_31864921 | 0.01 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr17_+_10593398 | 0.01 |
ENSDART00000168897
ENSDART00000193989 ENSDART00000191664 ENSDART00000167188 |
mapkbp1
|
mitogen-activated protein kinase binding protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.3 | GO:0036368 | cone photoresponse recovery(GO:0036368) |
| 0.2 | 1.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 1.2 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.1 | 0.5 | GO:0010872 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.7 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.1 | 0.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.5 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 1.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.4 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.1 | 0.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.5 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 2.2 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.1 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.2 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 3.0 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.4 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 2.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.6 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 1.7 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.2 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.4 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.6 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.3 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.1 | 0.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 2.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.4 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 2.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.7 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.6 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 1.3 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.3 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |