Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
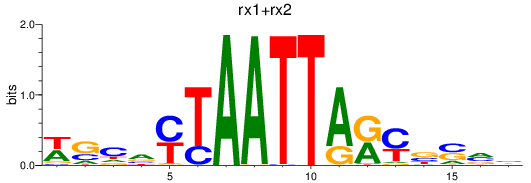
Results for rx1+rx2
Z-value: 0.36
Transcription factors associated with rx1+rx2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx2
|
ENSDARG00000040321 | retinal homeobox gene 2 |
|
rx1
|
ENSDARG00000071684 | retinal homeobox gene 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx1 | dr11_v1_chr22_-_5006119_5006119 | 0.49 | 3.4e-02 | Click! |
| rx2 | dr11_v1_chr2_-_55853943_55853943 | 0.38 | 1.1e-01 | Click! |
Activity profile of rx1+rx2 motif
Sorted Z-values of rx1+rx2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_306036 | 0.77 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr8_-_16697615 | 0.62 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr3_-_53533128 | 0.60 |
ENSDART00000183591
|
notch3
|
notch 3 |
| chr4_-_9891874 | 0.59 |
ENSDART00000067193
|
adm2a
|
adrenomedullin 2a |
| chr9_-_34937025 | 0.57 |
ENSDART00000137888
|
cdc16
|
cell division cycle 16 homolog (S. cerevisiae) |
| chr8_-_50888806 | 0.52 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr24_+_19415124 | 0.52 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr21_-_15200556 | 0.48 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr25_+_30131055 | 0.44 |
ENSDART00000152705
|
api5
|
apoptosis inhibitor 5 |
| chr11_-_21528056 | 0.44 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr11_-_39118882 | 0.42 |
ENSDART00000113185
ENSDART00000156526 |
ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr24_+_38301080 | 0.42 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr20_+_29209615 | 0.41 |
ENSDART00000062350
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr14_+_21820034 | 0.40 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr15_+_23799461 | 0.39 |
ENSDART00000154885
|
si:ch211-167j9.4
|
si:ch211-167j9.4 |
| chr25_+_11008419 | 0.38 |
ENSDART00000156589
|
mhc1lia
|
major histocompatibility complex class I LIA |
| chr1_+_21731382 | 0.37 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr23_+_43255328 | 0.36 |
ENSDART00000102712
|
tgm2a
|
transglutaminase 2, C polypeptide A |
| chr8_+_26059677 | 0.35 |
ENSDART00000009178
|
impdh2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr18_+_17827149 | 0.35 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr4_+_6032640 | 0.34 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr3_-_26017831 | 0.33 |
ENSDART00000179982
|
hmox1a
|
heme oxygenase 1a |
| chr8_+_18010978 | 0.32 |
ENSDART00000039887
ENSDART00000144532 |
ssbp3b
|
single stranded DNA binding protein 3b |
| chr10_-_2788668 | 0.32 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr14_-_4170654 | 0.31 |
ENSDART00000188347
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr16_+_23799622 | 0.30 |
ENSDART00000046922
|
rab13
|
RAB13, member RAS oncogene family |
| chr21_-_30082414 | 0.30 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr13_-_35808904 | 0.30 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr4_+_11723852 | 0.30 |
ENSDART00000028820
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr8_-_39822917 | 0.30 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr18_+_9171778 | 0.29 |
ENSDART00000101192
|
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr7_-_41746427 | 0.29 |
ENSDART00000174232
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr16_-_30434279 | 0.29 |
ENSDART00000018504
|
zgc:77086
|
zgc:77086 |
| chr9_+_43799829 | 0.29 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr16_-_27677930 | 0.28 |
ENSDART00000145991
|
tbrg4
|
transforming growth factor beta regulator 4 |
| chr11_+_16153207 | 0.28 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_+_24023653 | 0.28 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr20_+_36812368 | 0.28 |
ENSDART00000062931
|
abracl
|
ABRA C-terminal like |
| chr8_+_18010568 | 0.27 |
ENSDART00000121984
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr8_+_25079470 | 0.27 |
ENSDART00000000744
|
sypl2b
|
synaptophysin-like 2b |
| chr20_-_37813863 | 0.26 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr17_+_24318753 | 0.26 |
ENSDART00000064083
|
otx1
|
orthodenticle homeobox 1 |
| chr19_+_2631565 | 0.25 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr20_+_29209767 | 0.24 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr2_+_25315591 | 0.24 |
ENSDART00000161386
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr9_-_11676491 | 0.24 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr22_+_2260497 | 0.23 |
ENSDART00000142890
|
si:dkey-4c15.9
|
si:dkey-4c15.9 |
| chr5_-_12219572 | 0.23 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr8_+_29742237 | 0.23 |
ENSDART00000133955
ENSDART00000020621 |
mapk4
|
mitogen-activated protein kinase 4 |
| chr10_-_34002185 | 0.23 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr25_+_20715950 | 0.22 |
ENSDART00000180223
|
ergic2
|
ERGIC and golgi 2 |
| chr19_-_5669122 | 0.21 |
ENSDART00000112211
|
si:ch211-264f5.2
|
si:ch211-264f5.2 |
| chr8_+_18011522 | 0.21 |
ENSDART00000136756
|
ssbp3b
|
single stranded DNA binding protein 3b |
| chr6_+_36839509 | 0.20 |
ENSDART00000190605
ENSDART00000104160 |
zgc:110788
|
zgc:110788 |
| chr6_-_40922971 | 0.20 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr11_+_16152316 | 0.20 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr20_-_9095105 | 0.20 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr20_+_29209926 | 0.20 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr23_+_12134839 | 0.20 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr11_-_16152400 | 0.19 |
ENSDART00000123665
|
arpc4l
|
actin related protein 2/3 complex, subunit 4, like |
| chr8_+_29986265 | 0.18 |
ENSDART00000148258
|
ptch1
|
patched 1 |
| chr4_+_47636303 | 0.18 |
ENSDART00000167272
ENSDART00000166961 |
BX324142.1
|
|
| chr17_+_24821627 | 0.18 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr7_-_33960170 | 0.18 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr13_-_36798204 | 0.16 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr16_+_35379720 | 0.16 |
ENSDART00000170497
|
si:dkey-34d22.2
|
si:dkey-34d22.2 |
| chr17_+_20589553 | 0.15 |
ENSDART00000154447
|
si:ch73-288o11.4
|
si:ch73-288o11.4 |
| chr19_-_25149598 | 0.15 |
ENSDART00000162917
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_-_24332284 | 0.15 |
ENSDART00000163241
|
pimr142
|
Pim proto-oncogene, serine/threonine kinase, related 142 |
| chr5_-_25733745 | 0.15 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr8_+_28695914 | 0.14 |
ENSDART00000033386
|
ocstamp
|
osteoclast stimulatory transmembrane protein |
| chr6_-_7720332 | 0.14 |
ENSDART00000135945
|
rpsa
|
ribosomal protein SA |
| chr20_-_29864390 | 0.14 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr16_-_54971277 | 0.14 |
ENSDART00000113358
|
wdtc1
|
WD and tetratricopeptide repeats 1 |
| chr1_+_35985813 | 0.13 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr21_+_45223194 | 0.13 |
ENSDART00000150902
|
si:ch73-269m14.3
|
si:ch73-269m14.3 |
| chr7_+_6652967 | 0.13 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr8_-_24366726 | 0.13 |
ENSDART00000182351
ENSDART00000187099 ENSDART00000181534 |
CR450805.1
|
|
| chr5_-_52010122 | 0.13 |
ENSDART00000073627
ENSDART00000163898 ENSDART00000051003 |
cdk7
|
cyclin-dependent kinase 7 |
| chr6_-_55399214 | 0.13 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr17_-_9950911 | 0.13 |
ENSDART00000105117
|
sptssa
|
serine palmitoyltransferase, small subunit A |
| chr2_+_6255434 | 0.13 |
ENSDART00000139429
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_+_37640626 | 0.13 |
ENSDART00000008047
|
wdr24
|
WD repeat domain 24 |
| chr1_+_6161930 | 0.12 |
ENSDART00000037036
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr8_-_24502997 | 0.12 |
ENSDART00000184801
ENSDART00000179794 |
BX296549.1
|
|
| chr24_-_25144441 | 0.12 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr21_-_45363871 | 0.12 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr7_-_40630698 | 0.12 |
ENSDART00000134547
|
ube3c
|
ubiquitin protein ligase E3C |
| chr13_-_36663358 | 0.10 |
ENSDART00000085319
|
sos2
|
son of sevenless homolog 2 (Drosophila) |
| chr21_-_13661631 | 0.10 |
ENSDART00000184408
|
pnpla7a
|
patatin-like phospholipase domain containing 7a |
| chr8_-_53044300 | 0.09 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr6_+_40922572 | 0.09 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr18_-_40708537 | 0.09 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr16_+_13818743 | 0.09 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr1_+_6162090 | 0.08 |
ENSDART00000188295
|
cnppd1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr6_+_7421898 | 0.08 |
ENSDART00000043946
|
ccdc65
|
coiled-coil domain containing 65 |
| chr7_+_34453185 | 0.08 |
ENSDART00000173875
ENSDART00000173921 ENSDART00000173995 |
si:cabz01009626.1
|
si:cabz01009626.1 |
| chr21_-_2341937 | 0.08 |
ENSDART00000158459
|
zgc:193790
|
zgc:193790 |
| chr4_+_9400012 | 0.07 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr22_-_20924564 | 0.07 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr17_+_22587356 | 0.07 |
ENSDART00000157328
|
birc6
|
baculoviral IAP repeat containing 6 |
| chr9_+_21165484 | 0.07 |
ENSDART00000177286
|
si:rp71-68n21.9
|
si:rp71-68n21.9 |
| chr4_-_56954002 | 0.06 |
ENSDART00000160934
|
si:dkey-269o24.1
|
si:dkey-269o24.1 |
| chr17_-_53329704 | 0.06 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr17_-_49978986 | 0.05 |
ENSDART00000154728
|
col12a1a
|
collagen, type XII, alpha 1a |
| chr22_-_10055744 | 0.05 |
ENSDART00000143686
|
si:ch211-222k6.2
|
si:ch211-222k6.2 |
| chr8_-_53044089 | 0.05 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr21_-_43015383 | 0.05 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr13_+_4871886 | 0.05 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
| chr10_-_40421050 | 0.04 |
ENSDART00000141834
ENSDART00000164847 |
taar20w
|
trace amine associated receptor 20w |
| chr8_-_18010735 | 0.04 |
ENSDART00000125014
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr7_+_46368520 | 0.03 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr14_-_413273 | 0.03 |
ENSDART00000163976
ENSDART00000179907 |
FAT4
|
FAT atypical cadherin 4 |
| chr8_-_25034411 | 0.02 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr7_+_26173751 | 0.01 |
ENSDART00000065131
|
si:ch211-196f2.7
|
si:ch211-196f2.7 |
| chr22_+_23359369 | 0.01 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr22_+_5123479 | 0.01 |
ENSDART00000111822
ENSDART00000081910 |
loxl5a
|
lysyl oxidase-like 5a |
| chr25_+_34938317 | 0.00 |
ENSDART00000042678
|
vps4a
|
vacuolar protein sorting 4a homolog A (S. cerevisiae) |
| chr23_-_15216654 | 0.00 |
ENSDART00000131649
|
sulf2b
|
sulfatase 2b |
| chr19_+_42983613 | 0.00 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr21_-_33995710 | 0.00 |
ENSDART00000100508
ENSDART00000179622 |
ebf1b
|
early B cell factor 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of rx1+rx2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0048340 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 0.6 | GO:0098725 | somatic stem cell population maintenance(GO:0035019) symmetric cell division(GO:0098725) |
| 0.2 | 0.6 | GO:0016109 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.1 | 0.4 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.1 | 0.5 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.3 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.3 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.3 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.0 | 0.3 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.4 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.1 | GO:0045922 | negative regulation of fatty acid metabolic process(GO:0045922) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.5 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.3 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.0 | 0.4 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.4 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.4 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.0 | 0.5 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |