Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
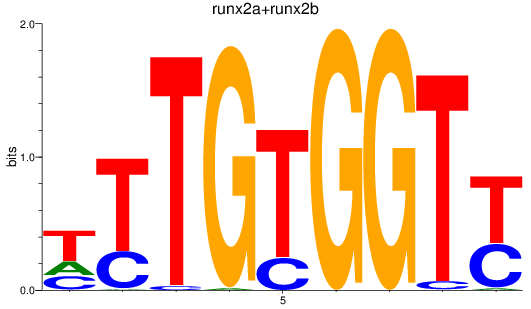
Results for runx2a+runx2b
Z-value: 1.16
Transcription factors associated with runx2a+runx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx2a
|
ENSDARG00000040261 | RUNX family transcription factor 2a |
|
runx2b
|
ENSDARG00000059233 | RUNX family transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx2a | dr11_v1_chr17_+_5351922_5351922 | -0.22 | 3.7e-01 | Click! |
| runx2b | dr11_v1_chr20_-_44055095_44055123 | -0.15 | 5.3e-01 | Click! |
Activity profile of runx2a+runx2b motif
Sorted Z-values of runx2a+runx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_6641223 | 2.95 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr10_-_25769334 | 2.83 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr9_-_22205682 | 2.67 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr11_+_25481046 | 2.63 |
ENSDART00000065940
|
opn1lw2
|
opsin 1 (cone pigments), long-wave-sensitive, 2 |
| chr21_+_7582036 | 2.53 |
ENSDART00000135485
ENSDART00000027268 |
otpa
|
orthopedia homeobox a |
| chr20_-_14054083 | 2.50 |
ENSDART00000009549
|
rhag
|
Rh associated glycoprotein |
| chr23_+_3538463 | 2.34 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr7_+_34231782 | 2.17 |
ENSDART00000173547
|
lctla
|
lactase-like a |
| chr7_+_25858380 | 1.94 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr8_+_25173317 | 1.91 |
ENSDART00000142006
|
gpr61
|
G protein-coupled receptor 61 |
| chr14_+_6159162 | 1.86 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr2_-_32768951 | 1.86 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr25_+_20216159 | 1.82 |
ENSDART00000048642
|
tnnt2d
|
troponin T2d, cardiac |
| chr16_-_16152199 | 1.78 |
ENSDART00000012718
|
fabp11b
|
fatty acid binding protein 11b |
| chr2_+_2223837 | 1.73 |
ENSDART00000101038
ENSDART00000129354 |
tmie
|
transmembrane inner ear |
| chr15_-_23376541 | 1.68 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr16_+_11724230 | 1.52 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr18_-_12052132 | 1.51 |
ENSDART00000074361
|
zgc:110789
|
zgc:110789 |
| chr17_+_26815021 | 1.42 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr10_-_15128771 | 1.37 |
ENSDART00000101261
|
spp1
|
secreted phosphoprotein 1 |
| chr19_+_30633453 | 1.30 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr19_+_37857936 | 1.27 |
ENSDART00000189289
|
nxph1
|
neurexophilin 1 |
| chr13_-_29424454 | 1.25 |
ENSDART00000026765
|
slc18a3a
|
solute carrier family 18 (vesicular acetylcholine transporter), member 3a |
| chr10_+_21797276 | 1.24 |
ENSDART00000169105
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr25_+_7982979 | 1.23 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr11_+_21053488 | 1.23 |
ENSDART00000189860
|
zgc:113307
|
zgc:113307 |
| chr1_+_15137901 | 1.22 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr1_-_26702930 | 1.20 |
ENSDART00000109297
ENSDART00000152389 |
foxe1
|
forkhead box E1 |
| chr25_-_7925269 | 1.17 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr3_+_54168007 | 1.17 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr19_+_32553874 | 1.16 |
ENSDART00000078197
|
heyl
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr13_+_27316934 | 1.15 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr2_-_22286828 | 1.14 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr23_+_8797143 | 1.13 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr16_+_50741154 | 1.13 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr6_-_39764995 | 1.10 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr7_+_46368520 | 1.09 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr25_-_7925019 | 1.09 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr16_+_11483811 | 1.08 |
ENSDART00000169012
ENSDART00000173042 |
grik5
|
glutamate receptor, ionotropic, kainate 5 |
| chr14_-_30967284 | 1.07 |
ENSDART00000149435
|
il2rgb
|
interleukin 2 receptor, gamma b |
| chr7_+_52811291 | 1.07 |
ENSDART00000192900
|
CT030160.1
|
|
| chr1_-_55810730 | 1.06 |
ENSDART00000100551
|
zgc:136908
|
zgc:136908 |
| chr6_+_2097690 | 1.04 |
ENSDART00000193770
|
tgm2b
|
transglutaminase 2b |
| chr11_-_12051805 | 1.01 |
ENSDART00000110117
ENSDART00000182744 |
socs7
|
suppressor of cytokine signaling 7 |
| chr16_+_31853919 | 1.01 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr7_-_35516251 | 0.99 |
ENSDART00000045628
|
irx6a
|
iroquois homeobox 6a |
| chr8_+_37489495 | 0.98 |
ENSDART00000141516
|
fmodb
|
fibromodulin b |
| chr5_-_39474235 | 0.96 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr5_+_23597144 | 0.96 |
ENSDART00000143929
ENSDART00000142420 ENSDART00000032909 |
kat5b
|
K(lysine) acetyltransferase 5b |
| chr23_-_27822920 | 0.95 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr3_-_28428198 | 0.95 |
ENSDART00000151546
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr6_-_43449013 | 0.93 |
ENSDART00000122423
|
eevs
|
2-epi-5-epi-valiolone synthase |
| chr4_-_8882572 | 0.93 |
ENSDART00000190060
|
mpped1
|
metallophosphoesterase domain containing 1 |
| chr3_+_15271943 | 0.92 |
ENSDART00000141752
|
asphd1
|
aspartate beta-hydroxylase domain containing 1 |
| chr2_-_985417 | 0.92 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr21_+_21309164 | 0.90 |
ENSDART00000132174
|
si:ch211-191j22.8
|
si:ch211-191j22.8 |
| chr17_-_46457622 | 0.90 |
ENSDART00000130215
|
TMEM179 (1 of many)
|
transmembrane protein 179 |
| chr7_-_38633867 | 0.90 |
ENSDART00000137424
|
c1qtnf4
|
C1q and TNF related 4 |
| chr7_-_38658411 | 0.90 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr18_-_10995410 | 0.88 |
ENSDART00000136751
|
tspan33b
|
tetraspanin 33b |
| chr15_-_12545683 | 0.87 |
ENSDART00000162807
|
scn2b
|
sodium channel, voltage-gated, type II, beta |
| chr25_+_19954576 | 0.87 |
ENSDART00000149335
|
kcna1a
|
potassium voltage-gated channel, shaker-related subfamily, member 1a |
| chr1_-_30689004 | 0.84 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr24_-_28648949 | 0.84 |
ENSDART00000180227
|
tecrl2a
|
trans-2,3-enoyl-CoA reductase-like 2a |
| chr23_-_36724575 | 0.83 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr21_-_32487061 | 0.82 |
ENSDART00000114359
ENSDART00000131591 ENSDART00000131477 |
si:dkeyp-72g9.4
|
si:dkeyp-72g9.4 |
| chr2_-_31936966 | 0.82 |
ENSDART00000169484
ENSDART00000192492 ENSDART00000027689 |
amph
|
amphiphysin |
| chr8_+_19356072 | 0.82 |
ENSDART00000063272
|
mpeg1.2
|
macrophage expressed 1, tandem duplicate 2 |
| chr19_+_40350468 | 0.82 |
ENSDART00000087444
|
hepacam2
|
HEPACAM family member 2 |
| chr6_+_48154954 | 0.82 |
ENSDART00000019706
|
phc2b
|
polyhomeotic homolog 2b (Drosophila) |
| chr17_+_24006792 | 0.81 |
ENSDART00000122415
|
si:ch211-63b16.4
|
si:ch211-63b16.4 |
| chr11_+_21076872 | 0.81 |
ENSDART00000155521
|
prelp
|
proline/arginine-rich end leucine-rich repeat protein |
| chr1_-_58064738 | 0.81 |
ENSDART00000073778
|
caspb
|
caspase b |
| chr6_-_12314475 | 0.81 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr12_-_15620090 | 0.80 |
ENSDART00000038032
|
acbd4
|
acyl-CoA binding domain containing 4 |
| chr15_+_37197494 | 0.80 |
ENSDART00000166203
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr10_-_20453995 | 0.79 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr13_+_31603988 | 0.77 |
ENSDART00000030646
|
six6a
|
SIX homeobox 6a |
| chr11_-_27057572 | 0.77 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr5_-_68876211 | 0.77 |
ENSDART00000097254
|
sftpbb
|
surfactant protein Bb |
| chr13_+_10232695 | 0.76 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr7_-_35515931 | 0.76 |
ENSDART00000193324
|
irx6a
|
iroquois homeobox 6a |
| chr14_-_17068511 | 0.76 |
ENSDART00000163766
|
phox2bb
|
paired-like homeobox 2bb |
| chr22_-_10459880 | 0.74 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr7_-_18656069 | 0.73 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr14_-_4076480 | 0.72 |
ENSDART00000059231
|
enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr22_+_21317597 | 0.72 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr23_-_29376859 | 0.71 |
ENSDART00000146411
|
sst6
|
somatostatin 6 |
| chr10_-_7974155 | 0.71 |
ENSDART00000147368
ENSDART00000075524 |
osbp2
|
oxysterol binding protein 2 |
| chr23_+_35847538 | 0.71 |
ENSDART00000143935
|
rarga
|
retinoic acid receptor gamma a |
| chr14_-_17068712 | 0.71 |
ENSDART00000170277
|
phox2bb
|
paired-like homeobox 2bb |
| chr16_+_32995882 | 0.71 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr24_+_32472155 | 0.70 |
ENSDART00000098859
|
neurod6a
|
neuronal differentiation 6a |
| chr16_-_17200120 | 0.70 |
ENSDART00000147739
|
gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr4_-_15420452 | 0.69 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr3_+_14339728 | 0.69 |
ENSDART00000184127
|
plppr2a
|
phospholipid phosphatase related 2a |
| chr7_+_13382852 | 0.68 |
ENSDART00000166318
|
dagla
|
diacylglycerol lipase, alpha |
| chr22_-_33679277 | 0.68 |
ENSDART00000169948
|
FO904977.1
|
|
| chr20_+_5564042 | 0.68 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr3_-_36440705 | 0.67 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr9_-_23118350 | 0.67 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr2_-_50298337 | 0.66 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr15_-_29586747 | 0.66 |
ENSDART00000076749
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr1_-_51465730 | 0.65 |
ENSDART00000074284
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr11_-_12051502 | 0.65 |
ENSDART00000183462
|
socs7
|
suppressor of cytokine signaling 7 |
| chr13_+_32144370 | 0.64 |
ENSDART00000020270
|
osr1
|
odd-skipped related transciption factor 1 |
| chr10_+_21796477 | 0.64 |
ENSDART00000176255
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr25_-_18249751 | 0.64 |
ENSDART00000153950
|
si:dkey-106n21.1
|
si:dkey-106n21.1 |
| chr7_-_61845282 | 0.63 |
ENSDART00000182586
|
HTRA3
|
HtrA serine peptidase 3 |
| chr9_-_28867562 | 0.63 |
ENSDART00000189597
ENSDART00000060321 |
zgc:91818
|
zgc:91818 |
| chr23_-_44207349 | 0.63 |
ENSDART00000186276
|
zgc:158659
|
zgc:158659 |
| chr10_-_8053385 | 0.63 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr8_-_29713595 | 0.62 |
ENSDART00000131988
ENSDART00000077637 |
mpeg1.1
|
macrophage expressed 1, tandem duplicate 1 |
| chr13_+_27316632 | 0.62 |
ENSDART00000016121
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr15_-_38154616 | 0.62 |
ENSDART00000099392
|
irgq2
|
immunity-related GTPase family, q2 |
| chr16_-_16619854 | 0.62 |
ENSDART00000150512
ENSDART00000191306 ENSDART00000181773 ENSDART00000183231 |
cyp21a2
|
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr25_+_20119466 | 0.62 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr12_+_26467847 | 0.62 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr4_-_2525916 | 0.61 |
ENSDART00000134123
ENSDART00000132581 ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr19_+_25504645 | 0.61 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr25_-_3470910 | 0.61 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr8_-_46926204 | 0.60 |
ENSDART00000188758
ENSDART00000143321 |
hes2.1
|
hes family bHLH transcription factor 2, tandem duplicate 1 |
| chr11_-_12051283 | 0.60 |
ENSDART00000170516
|
socs7
|
suppressor of cytokine signaling 7 |
| chr19_+_40856807 | 0.60 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr6_+_9867426 | 0.60 |
ENSDART00000151749
|
MPP4 (1 of many)
|
si:ch211-222n4.6 |
| chr10_+_15255198 | 0.59 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr15_-_2184638 | 0.59 |
ENSDART00000135460
|
shox2
|
short stature homeobox 2 |
| chr20_+_18225329 | 0.59 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr8_-_17067364 | 0.59 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr16_-_39900665 | 0.59 |
ENSDART00000136719
|
rbms3
|
RNA binding motif, single stranded interacting protein |
| chr14_-_2163454 | 0.59 |
ENSDART00000160123
ENSDART00000169653 |
pcdh2ab9
pcdh2ac
|
protocadherin 2 alpha b 9 protocadherin 2 alpha c |
| chr12_-_25612170 | 0.58 |
ENSDART00000077155
|
six2b
|
SIX homeobox 2b |
| chr5_-_52277643 | 0.58 |
ENSDART00000010757
|
rgmb
|
repulsive guidance molecule family member b |
| chr19_+_8144556 | 0.58 |
ENSDART00000027274
ENSDART00000147218 |
efna3a
|
ephrin-A3a |
| chr3_-_5664123 | 0.58 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr13_-_23095006 | 0.58 |
ENSDART00000089242
|
kif1bp
|
kif1 binding protein |
| chr5_+_44190974 | 0.58 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr10_+_25355308 | 0.58 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr2_-_9748039 | 0.58 |
ENSDART00000134870
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr10_+_15255012 | 0.57 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr10_-_43771447 | 0.57 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr19_-_10656667 | 0.57 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr2_-_51087077 | 0.56 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr1_+_41854298 | 0.56 |
ENSDART00000192672
|
smox
|
spermine oxidase |
| chr19_+_40856534 | 0.55 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr15_-_29573267 | 0.55 |
ENSDART00000099947
|
samsn1a
|
SAM domain, SH3 domain and nuclear localisation signals 1a |
| chr14_+_30491890 | 0.55 |
ENSDART00000131174
|
fgf20b
|
fibroblast growth factor 20b |
| chr18_+_45573251 | 0.54 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr1_-_16394814 | 0.54 |
ENSDART00000013024
|
fgf20a
|
fibroblast growth factor 20a |
| chr5_+_61467618 | 0.53 |
ENSDART00000074073
ENSDART00000182094 |
alkbh4
|
alkB homolog 4, lysine demthylase |
| chr13_+_25433774 | 0.53 |
ENSDART00000141255
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr4_+_2620751 | 0.53 |
ENSDART00000013924
|
gpr22a
|
G protein-coupled receptor 22a |
| chr22_-_5323482 | 0.53 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr1_+_46509176 | 0.52 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr23_+_28582865 | 0.52 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr19_-_5812319 | 0.52 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr24_+_11334733 | 0.52 |
ENSDART00000147552
ENSDART00000143171 |
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr5_+_57658898 | 0.52 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr21_+_33503835 | 0.51 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr15_-_24869826 | 0.51 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr24_-_31452875 | 0.51 |
ENSDART00000187381
ENSDART00000185128 |
cngb3.2
|
cyclic nucleotide gated channel beta 3, tandem duplicate 2 |
| chr13_-_10261383 | 0.51 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr6_+_38381957 | 0.50 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr25_-_7974494 | 0.50 |
ENSDART00000171446
|
hal
|
histidine ammonia-lyase |
| chr7_+_36898850 | 0.50 |
ENSDART00000113342
|
tox3
|
TOX high mobility group box family member 3 |
| chr5_-_33039670 | 0.49 |
ENSDART00000141361
|
glipr2
|
GLI pathogenesis-related 2 |
| chr8_-_44868020 | 0.49 |
ENSDART00000142712
|
cacna1fa
|
calcium channel, voltage-dependent, L type, alpha 1F subunit a |
| chr3_-_50954607 | 0.49 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr17_-_40397752 | 0.48 |
ENSDART00000178483
|
BX548062.1
|
|
| chr4_-_10835620 | 0.48 |
ENSDART00000150739
|
ppfibp1a
|
PTPRF interacting protein, binding protein 1a (liprin beta 1) |
| chr3_+_58379450 | 0.48 |
ENSDART00000155759
|
sdr42e2
|
short chain dehydrogenase/reductase family 42E, member 2 |
| chr11_+_14287427 | 0.48 |
ENSDART00000180903
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr18_+_45573416 | 0.48 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr1_+_23408622 | 0.48 |
ENSDART00000140706
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr3_+_37197686 | 0.47 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr16_-_54455573 | 0.47 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr20_+_33904258 | 0.47 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr6_+_13806466 | 0.47 |
ENSDART00000043522
|
tmem198b
|
transmembrane protein 198b |
| chr9_-_40765868 | 0.46 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr6_+_52804267 | 0.46 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr13_+_22675802 | 0.46 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr7_+_12976173 | 0.46 |
ENSDART00000193150
ENSDART00000149518 |
slc25a22b
|
solute carrier family 25 member 22b |
| chr15_+_17446796 | 0.46 |
ENSDART00000157189
|
snx19b
|
sorting nexin 19b |
| chr5_+_65991152 | 0.45 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr9_-_25425381 | 0.45 |
ENSDART00000129522
|
acvr2aa
|
activin A receptor type 2Aa |
| chr11_+_24815667 | 0.45 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr10_-_17103651 | 0.45 |
ENSDART00000108959
|
RNF208
|
ring finger protein 208 |
| chr20_+_20499869 | 0.45 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr20_+_43083745 | 0.45 |
ENSDART00000139014
ENSDART00000153438 |
moxd1l
|
monooxygenase, DBH-like 1, like |
| chr20_+_32406011 | 0.44 |
ENSDART00000018640
ENSDART00000137910 |
snx3
|
sorting nexin 3 |
| chr25_-_25434479 | 0.44 |
ENSDART00000171589
|
hrasa
|
v-Ha-ras Harvey rat sarcoma viral oncogene homolog a |
| chr3_+_50310684 | 0.44 |
ENSDART00000112152
|
gas7a
|
growth arrest-specific 7a |
| chr7_-_33248773 | 0.44 |
ENSDART00000052389
|
itga11a
|
integrin, alpha 11a |
| chr19_+_2546775 | 0.43 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr22_-_34937455 | 0.42 |
ENSDART00000169217
ENSDART00000188330 ENSDART00000165142 |
slit1b
|
slit homolog 1b (Drosophila) |
| chr24_-_18877118 | 0.42 |
ENSDART00000092783
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr21_+_15824182 | 0.42 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr12_+_38774860 | 0.42 |
ENSDART00000130371
|
kif19
|
kinesin family member 19 |
| chr14_-_6727717 | 0.42 |
ENSDART00000166979
|
si:dkeyp-44a8.4
|
si:dkeyp-44a8.4 |
| chr10_+_17088261 | 0.42 |
ENSDART00000132103
|
si:dkey-106l3.7
|
si:dkey-106l3.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx2a+runx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.5 | GO:0015840 | urea transport(GO:0015840) |
| 0.5 | 1.4 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.5 | 2.7 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.4 | 1.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.4 | 1.5 | GO:0061549 | sympathetic ganglion development(GO:0061549) |
| 0.4 | 1.1 | GO:0051230 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.3 | 3.0 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 1.9 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 0.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 0.7 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.2 | 0.6 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.2 | 2.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.2 | 0.7 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.2 | 0.5 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.4 | GO:0042421 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.1 | 0.7 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.1 | 0.4 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.1 | 0.4 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.1 | 0.7 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.7 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.6 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.1 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.9 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 1.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.5 | GO:0001659 | temperature homeostasis(GO:0001659) |
| 0.1 | 0.6 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.3 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 1.2 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.1 | 0.4 | GO:0045056 | transcytosis(GO:0045056) |
| 0.1 | 0.4 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.1 | 0.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.2 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.1 | 1.0 | GO:0098868 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.1 | 0.3 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.1 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 2.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.5 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.3 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.1 | 0.3 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.1 | 0.5 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.1 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.5 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.5 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.5 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.3 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.0 | 0.4 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.1 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0055057 | neuroblast division(GO:0055057) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 2.3 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.0 | 1.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.5 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.7 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.0 | 0.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 1.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.3 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.2 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 1.1 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.3 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.5 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.8 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.5 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 1.2 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 1.2 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 2.5 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.6 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.1 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 1.2 | GO:0060348 | bone development(GO:0060348) |
| 0.0 | 2.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 1.3 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.3 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 1.4 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.2 | GO:0099633 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.7 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0098900 | regulation of action potential(GO:0098900) |
| 0.0 | 0.6 | GO:0021761 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.3 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.5 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 3.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.0 | 0.1 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.9 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.0 | GO:0002836 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 1.3 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 1.4 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.8 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.2 | 1.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 1.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.9 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 1.2 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.7 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 1.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.6 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 1.0 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.8 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 1.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.9 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.3 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 1.0 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.4 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.3 | 3.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 1.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.2 | 1.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.2 | 1.9 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.2 | 2.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.2 | 0.7 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.2 | 1.8 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.2 | 0.9 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.4 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.1 | 0.4 | GO:0031530 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.1 | 3.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.6 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.5 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.1 | 0.3 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.1 | 0.7 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.6 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 1.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.5 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.5 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.9 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.5 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.6 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.7 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.4 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 2.8 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.9 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.1 | 1.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.7 | GO:0005223 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.1 | 0.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 1.4 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 1.0 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.6 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.7 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 2.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:1902387 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.7 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.6 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0015172 | L-glutamate transmembrane transporter activity(GO:0005313) acidic amino acid transmembrane transporter activity(GO:0015172) L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.1 | GO:0033745 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.4 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.9 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 2.5 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 1.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.7 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.1 | 1.2 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 0.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 1.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 3.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.1 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.7 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 1.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 0.9 | REACTOME HIGHLY CALCIUM PERMEABLE POSTSYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Highly calcium permeable postsynaptic nicotinic acetylcholine receptors |
| 0.1 | 1.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.1 | 1.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 0.8 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.6 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.6 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 1.2 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |