Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
Results for runx1
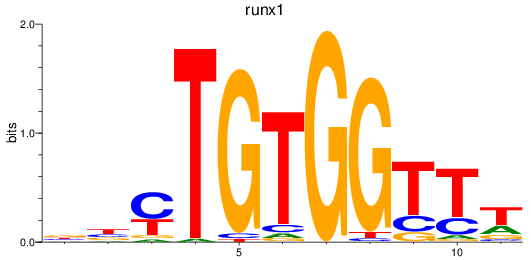
Z-value: 1.23
Transcription factors associated with runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx1
|
ENSDARG00000087646 | RUNX family transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx1 | dr11_v1_chr1_-_1402303_1402303 | -0.26 | 2.9e-01 | Click! |
Activity profile of runx1 motif
Sorted Z-values of runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_77973876 | 3.60 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr7_-_17337233 | 3.17 |
ENSDART00000050236
ENSDART00000102141 |
nitr8
|
novel immune-type receptor 8 |
| chr20_+_98179 | 3.11 |
ENSDART00000022725
|
si:ch1073-155h21.1
|
si:ch1073-155h21.1 |
| chr3_+_28576173 | 2.69 |
ENSDART00000151189
|
sept12
|
septin 12 |
| chr24_-_21921262 | 2.47 |
ENSDART00000186061
ENSDART00000187846 |
tagln3b
|
transgelin 3b |
| chr13_-_39947335 | 2.32 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr3_-_19367081 | 2.30 |
ENSDART00000191369
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr24_+_13635108 | 2.29 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr13_-_6081803 | 2.27 |
ENSDART00000099224
|
dld
|
deltaD |
| chr20_+_14941264 | 2.13 |
ENSDART00000080224
|
itpa
|
inosine triphosphatase (nucleoside triphosphate pyrophosphatase) |
| chr9_-_27410597 | 2.11 |
ENSDART00000135652
ENSDART00000042297 |
kdelc1
|
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr1_-_59571758 | 2.08 |
ENSDART00000193546
ENSDART00000167087 |
wfikkn1
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 1 |
| chr14_-_22118055 | 2.06 |
ENSDART00000179892
|
si:dkey-6i22.5
|
si:dkey-6i22.5 |
| chr7_-_48665305 | 2.01 |
ENSDART00000190507
|
cdkn1ca
|
cyclin-dependent kinase inhibitor 1Ca |
| chr17_+_24687338 | 1.98 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr7_-_24995631 | 1.97 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr3_-_58650057 | 1.96 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr4_+_63088692 | 1.94 |
ENSDART00000168872
|
zgc:173714
|
zgc:173714 |
| chr5_+_36513605 | 1.90 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr9_-_7390388 | 1.89 |
ENSDART00000132392
|
slc23a3
|
solute carrier family 23, member 3 |
| chr13_+_24263049 | 1.87 |
ENSDART00000135992
ENSDART00000088005 |
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr23_+_42362337 | 1.83 |
ENSDART00000164249
|
cyp2aa4
|
cytochrome P450, family 2, subfamily AA, polypeptide 4 |
| chr17_-_14817458 | 1.79 |
ENSDART00000155907
ENSDART00000156476 |
nid2a
|
nidogen 2a (osteonidogen) |
| chr12_+_6065661 | 1.79 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr3_+_54761569 | 1.77 |
ENSDART00000135913
ENSDART00000180983 |
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr14_-_41478265 | 1.77 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr24_+_19518570 | 1.74 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr23_+_5104743 | 1.71 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr4_-_38877924 | 1.71 |
ENSDART00000177741
|
znf1127
|
zinc finger protein 1127 |
| chr4_+_23117557 | 1.69 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr13_-_43108693 | 1.68 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr3_+_14641962 | 1.66 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr12_-_48312647 | 1.65 |
ENSDART00000114415
|
ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr24_+_31361407 | 1.65 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr18_+_22109379 | 1.64 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr13_+_10023256 | 1.62 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr7_-_60831082 | 1.59 |
ENSDART00000073654
ENSDART00000136999 |
pcxb
|
pyruvate carboxylase b |
| chr6_-_27108844 | 1.58 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr25_-_12996439 | 1.57 |
ENSDART00000171167
|
ccl39.4
|
chemokine (C-C motif) ligand 39, duplicate 4 |
| chr16_+_1383914 | 1.57 |
ENSDART00000185089
|
cers2b
|
ceramide synthase 2b |
| chr14_-_26498196 | 1.56 |
ENSDART00000054175
ENSDART00000145625 ENSDART00000183347 ENSDART00000191084 ENSDART00000191143 |
smad5
|
SMAD family member 5 |
| chr8_-_22964486 | 1.53 |
ENSDART00000112381
|
emilin3a
|
elastin microfibril interfacer 3a |
| chr12_-_13905307 | 1.51 |
ENSDART00000152400
|
dbf4b
|
DBF4 zinc finger B |
| chr4_+_13909398 | 1.51 |
ENSDART00000187959
ENSDART00000184926 |
pphln1
|
periphilin 1 |
| chr17_+_691453 | 1.50 |
ENSDART00000159271
|
fancm
|
Fanconi anemia, complementation group M |
| chr8_-_32803227 | 1.50 |
ENSDART00000110079
|
zgc:194839
|
zgc:194839 |
| chr19_-_35439237 | 1.50 |
ENSDART00000145883
|
anln
|
anillin, actin binding protein |
| chr22_+_21255860 | 1.49 |
ENSDART00000134893
|
cpamd8
|
C3 and PZP-like, alpha-2-macroglobulin domain containing 8 |
| chr9_-_40683722 | 1.49 |
ENSDART00000141979
ENSDART00000181228 |
bard1
|
BRCA1 associated RING domain 1 |
| chr1_+_604127 | 1.45 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr6_+_45932276 | 1.44 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr11_-_39202915 | 1.43 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr8_-_37249813 | 1.42 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr8_+_21353878 | 1.42 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr21_-_26495700 | 1.40 |
ENSDART00000109379
|
cd248b
|
CD248 molecule, endosialin b |
| chr24_-_7826489 | 1.40 |
ENSDART00000112777
|
si:dkey-197c15.6
|
si:dkey-197c15.6 |
| chr3_-_32362872 | 1.40 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr3_-_19368435 | 1.36 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr13_-_24263682 | 1.36 |
ENSDART00000176800
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr5_-_41860556 | 1.35 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr20_+_35282682 | 1.35 |
ENSDART00000187199
|
fam49a
|
family with sequence similarity 49, member A |
| chr17_+_5976683 | 1.34 |
ENSDART00000110276
|
zgc:194275
|
zgc:194275 |
| chr15_+_24572926 | 1.34 |
ENSDART00000155636
ENSDART00000187800 |
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr14_+_97017 | 1.34 |
ENSDART00000159300
ENSDART00000169523 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr19_-_18152407 | 1.34 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr1_-_45213565 | 1.31 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr18_-_2639351 | 1.31 |
ENSDART00000168106
|
relt
|
RELT, TNF receptor |
| chr15_+_37412883 | 1.31 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr3_-_27072179 | 1.31 |
ENSDART00000156556
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr21_-_14826066 | 1.29 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr14_-_31619408 | 1.27 |
ENSDART00000173277
|
mmgt1
|
membrane magnesium transporter 1 |
| chr22_+_9922301 | 1.26 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr19_+_19786117 | 1.26 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr21_-_26071142 | 1.25 |
ENSDART00000004740
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr9_+_38457806 | 1.25 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr10_+_24692076 | 1.25 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr5_+_30520249 | 1.24 |
ENSDART00000013431
|
hmbsa
|
hydroxymethylbilane synthase a |
| chr12_+_10443785 | 1.24 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr8_+_26432677 | 1.24 |
ENSDART00000078369
ENSDART00000131925 |
zgc:136971
|
zgc:136971 |
| chr21_+_34929598 | 1.23 |
ENSDART00000135806
|
si:dkey-71d15.2
|
si:dkey-71d15.2 |
| chr1_-_39859626 | 1.23 |
ENSDART00000053763
|
dctd
|
dCMP deaminase |
| chr9_-_24218059 | 1.22 |
ENSDART00000007914
|
nabp1a
|
nucleic acid binding protein 1a |
| chr6_+_40832635 | 1.21 |
ENSDART00000011931
|
ruvbl1
|
RuvB-like AAA ATPase 1 |
| chr11_+_25403561 | 1.19 |
ENSDART00000089120
|
adnpa
|
activity-dependent neuroprotector homeobox a |
| chr6_-_18960105 | 1.18 |
ENSDART00000185278
ENSDART00000162968 |
sept9b
|
septin 9b |
| chr19_+_27859546 | 1.17 |
ENSDART00000161908
|
nsun2
|
NOP2/Sun RNA methyltransferase family, member 2 |
| chr3_+_22335030 | 1.16 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr3_+_52078798 | 1.16 |
ENSDART00000156882
|
si:dkey-88e12.3
|
si:dkey-88e12.3 |
| chr9_-_24218367 | 1.16 |
ENSDART00000135356
|
nabp1a
|
nucleic acid binding protein 1a |
| chr25_-_14637660 | 1.15 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr23_+_36653376 | 1.15 |
ENSDART00000053189
|
gpr182
|
G protein-coupled receptor 182 |
| chr21_-_35324091 | 1.15 |
ENSDART00000185042
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr8_-_16697615 | 1.15 |
ENSDART00000187929
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr18_-_21929426 | 1.14 |
ENSDART00000132034
ENSDART00000079050 |
nutf2
|
nuclear transport factor 2 |
| chr2_+_20793982 | 1.14 |
ENSDART00000014785
|
prg4a
|
proteoglycan 4a |
| chr7_-_3894831 | 1.14 |
ENSDART00000172921
|
si:dkey-88n24.11
|
si:dkey-88n24.11 |
| chr13_+_11829072 | 1.14 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr10_+_19039507 | 1.14 |
ENSDART00000193538
ENSDART00000111952 ENSDART00000180093 |
igsf9a
|
immunoglobulin superfamily, member 9a |
| chr22_+_18156000 | 1.13 |
ENSDART00000143483
ENSDART00000136133 |
nr2c2ap
|
nuclear receptor 2C2-associated protein |
| chr8_-_47844456 | 1.12 |
ENSDART00000145429
|
si:dkeyp-104h9.5
|
si:dkeyp-104h9.5 |
| chr2_-_30055432 | 1.12 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr3_-_8285123 | 1.12 |
ENSDART00000158699
ENSDART00000138588 |
trim35-9
|
tripartite motif containing 35-9 |
| chr17_+_51744450 | 1.11 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr4_-_72295710 | 1.11 |
ENSDART00000182983
|
si:cabz01071911.3
|
si:cabz01071911.3 |
| chr14_+_26224541 | 1.11 |
ENSDART00000128971
|
gm2a
|
GM2 ganglioside activator |
| chr14_+_989733 | 1.10 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr7_+_15329819 | 1.10 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr20_+_30378803 | 1.09 |
ENSDART00000148242
ENSDART00000169140 ENSDART00000062441 |
rnaseh1
|
ribonuclease H1 |
| chr22_+_9862243 | 1.09 |
ENSDART00000105942
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr12_-_26851726 | 1.08 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr24_-_9989634 | 1.08 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr11_+_37720756 | 1.07 |
ENSDART00000173337
|
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr9_+_30112423 | 1.07 |
ENSDART00000112398
ENSDART00000013591 |
tfg
|
trk-fused gene |
| chr10_+_32663180 | 1.07 |
ENSDART00000138460
|
rad51c
|
RAD51 paralog C |
| chr6_+_29386728 | 1.07 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr2_-_16565690 | 1.07 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr9_+_38458193 | 1.07 |
ENSDART00000008053
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr17_-_28097760 | 1.06 |
ENSDART00000149861
|
kdm1a
|
lysine (K)-specific demethylase 1a |
| chr5_+_26138313 | 1.06 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr11_+_24815667 | 1.06 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr14_-_26436951 | 1.06 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr23_+_10347851 | 1.05 |
ENSDART00000127667
|
krt18
|
keratin 18 |
| chr12_-_3978306 | 1.05 |
ENSDART00000149473
ENSDART00000114857 |
ppp4cb
|
protein phosphatase 4, catalytic subunit b |
| chr1_+_39859782 | 1.05 |
ENSDART00000149984
|
irf2a
|
interferon regulatory factor 2a |
| chr11_+_6116503 | 1.05 |
ENSDART00000176170
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr16_+_32082547 | 1.04 |
ENSDART00000190122
|
prpf31
|
PRP31 pre-mRNA processing factor 31 homolog (yeast) |
| chr18_-_22735002 | 1.04 |
ENSDART00000023721
|
nudt21
|
nudix hydrolase 21 |
| chr2_+_23351454 | 1.04 |
ENSDART00000017034
ENSDART00000154645 |
rnf2
|
ring finger protein 2 |
| chr7_-_26262978 | 1.04 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr3_-_79366 | 1.04 |
ENSDART00000114289
|
zgc:165518
|
zgc:165518 |
| chr24_-_13349464 | 1.04 |
ENSDART00000134482
ENSDART00000139212 |
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr19_-_22328154 | 1.03 |
ENSDART00000090464
|
si:ch73-196l6.5
|
si:ch73-196l6.5 |
| chr3_-_7948799 | 1.03 |
ENSDART00000163714
|
trim35-24
|
tripartite motif containing 35-24 |
| chr14_+_38786298 | 1.03 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr11_+_37720552 | 1.03 |
ENSDART00000050296
ENSDART00000172867 |
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr25_-_16076257 | 1.02 |
ENSDART00000140780
|
ovch2
|
ovochymase 2 |
| chr16_+_13860299 | 1.02 |
ENSDART00000121998
|
grwd1
|
glutamate-rich WD repeat containing 1 |
| chr20_-_2725930 | 1.01 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr5_+_36899691 | 1.01 |
ENSDART00000132322
|
hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr21_-_2958422 | 1.01 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr22_-_38934989 | 1.01 |
ENSDART00000008365
|
ncbp2
|
nuclear cap binding protein subunit 2 |
| chr4_-_56898328 | 1.01 |
ENSDART00000169189
|
si:dkey-269o24.6
|
si:dkey-269o24.6 |
| chr9_-_14504834 | 1.00 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr15_+_30310843 | 0.99 |
ENSDART00000112784
|
lyrm9
|
LYR motif containing 9 |
| chr5_-_22600881 | 0.99 |
ENSDART00000176442
|
nono
|
non-POU domain containing, octamer-binding |
| chr5_+_63302660 | 0.99 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr2_+_30481125 | 0.99 |
ENSDART00000125933
|
fam173b
|
family with sequence similarity 173, member B |
| chr20_+_13883131 | 0.99 |
ENSDART00000003248
ENSDART00000152611 |
nek2
|
NIMA-related kinase 2 |
| chr2_+_3044992 | 0.98 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
| chr19_-_43657468 | 0.98 |
ENSDART00000150940
|
si:ch211-193k19.2
|
si:ch211-193k19.2 |
| chr25_-_12824656 | 0.98 |
ENSDART00000171801
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr13_+_48358467 | 0.97 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr7_+_17543487 | 0.97 |
ENSDART00000126652
|
CU672228.2
|
Danio rerio novel immune-type receptor 1l (nitr1l), mRNA. |
| chr4_+_15983541 | 0.96 |
ENSDART00000043660
|
copg2
|
coatomer protein complex, subunit gamma 2 |
| chr4_+_51347000 | 0.96 |
ENSDART00000161859
|
si:dkeyp-82b4.3
|
si:dkeyp-82b4.3 |
| chr12_+_33975065 | 0.96 |
ENSDART00000036649
|
sfxn2
|
sideroflexin 2 |
| chr15_-_2734560 | 0.95 |
ENSDART00000153853
|
ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr6_+_17789481 | 0.95 |
ENSDART00000190768
|
CR376861.1
|
|
| chr22_+_9862466 | 0.95 |
ENSDART00000146864
|
si:dkey-253d23.3
|
si:dkey-253d23.3 |
| chr21_-_26715270 | 0.95 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr6_-_2162446 | 0.95 |
ENSDART00000171265
|
tgm5l
|
transglutaminase 5, like |
| chr12_+_13905286 | 0.95 |
ENSDART00000147186
|
fkbp10b
|
FK506 binding protein 10b |
| chr12_-_20616160 | 0.95 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr12_-_3077395 | 0.95 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr20_-_9674877 | 0.95 |
ENSDART00000134097
|
nid2b
|
nidogen 2b (osteonidogen) |
| chr13_+_18533005 | 0.95 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr2_-_21786826 | 0.94 |
ENSDART00000016208
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr2_+_24374669 | 0.94 |
ENSDART00000133818
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr2_-_48539673 | 0.93 |
ENSDART00000168202
|
CR391991.4
|
|
| chr25_-_12824939 | 0.93 |
ENSDART00000169126
|
uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr8_+_19489854 | 0.93 |
ENSDART00000184671
ENSDART00000011258 |
npl
|
N-acetylneuraminate pyruvate lyase (dihydrodipicolinate synthase) |
| chr19_-_3874986 | 0.92 |
ENSDART00000161830
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr5_+_61475451 | 0.92 |
ENSDART00000163444
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr14_+_21820034 | 0.92 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr1_+_594584 | 0.92 |
ENSDART00000135944
|
jam2a
|
junctional adhesion molecule 2a |
| chr3_+_15809098 | 0.91 |
ENSDART00000183023
|
phospho1
|
phosphatase, orphan 1 |
| chr6_-_10725847 | 0.91 |
ENSDART00000184567
|
sp3b
|
Sp3b transcription factor |
| chr2_-_1227221 | 0.91 |
ENSDART00000130897
|
ABCF3
|
ATP binding cassette subfamily F member 3 |
| chr1_-_9109699 | 0.91 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr17_+_15534815 | 0.91 |
ENSDART00000159426
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr5_+_18925367 | 0.90 |
ENSDART00000051621
|
pgam5
|
PGAM family member 5, serine/threonine protein phosphatase, mitochondrial |
| chr13_+_36923052 | 0.90 |
ENSDART00000026313
|
tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr4_-_4261673 | 0.89 |
ENSDART00000150694
|
cd9b
|
CD9 molecule b |
| chr8_+_3434146 | 0.89 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr5_+_28497956 | 0.89 |
ENSDART00000191935
|
nfr
|
notochord formation related |
| chr17_-_9950911 | 0.89 |
ENSDART00000105117
|
sptssa
|
serine palmitoyltransferase, small subunit A |
| chr12_+_18782821 | 0.88 |
ENSDART00000152918
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr2_-_27619954 | 0.88 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr17_+_34244345 | 0.87 |
ENSDART00000006058
|
eif2s1a
|
eukaryotic translation initiation factor 2, subunit 1 alpha a |
| chr16_-_12060488 | 0.87 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr19_+_34742706 | 0.87 |
ENSDART00000103276
|
fam206a
|
family with sequence similarity 206, member A |
| chr6_-_34838397 | 0.87 |
ENSDART00000060169
ENSDART00000169605 |
mier1a
|
mesoderm induction early response 1a, transcriptional regulator |
| chr1_+_6226135 | 0.87 |
ENSDART00000108816
|
fastkd2
|
FAST kinase domains 2 |
| chr8_-_14091886 | 0.86 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr16_-_43679611 | 0.86 |
ENSDART00000123585
|
LO017721.1
|
|
| chr1_-_17711636 | 0.86 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr14_+_34501245 | 0.86 |
ENSDART00000131424
|
lcp2b
|
lymphocyte cytosolic protein 2b |
| chr2_+_30480907 | 0.86 |
ENSDART00000041378
ENSDART00000138863 |
fam173b
|
family with sequence similarity 173, member B |
| chr3_+_43774369 | 0.86 |
ENSDART00000157964
|
zc3h7a
|
zinc finger CCCH-type containing 7A |
| chr11_-_37720024 | 0.86 |
ENSDART00000023388
|
yod1
|
YOD1 deubiquitinase |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 1.0 | GO:0032241 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.9 | 3.6 | GO:0032208 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.8 | 2.3 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.7 | 2.8 | GO:0009221 | pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 0.6 | 0.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.6 | 1.7 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.4 | 2.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.4 | 1.5 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.4 | 2.5 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.4 | 1.4 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.3 | 1.0 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.3 | 1.0 | GO:0006290 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.3 | 2.3 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.3 | 2.2 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.3 | 3.1 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.3 | 1.8 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.5 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.3 | 2.0 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.3 | 1.1 | GO:0016108 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.3 | 1.1 | GO:1904590 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.3 | 0.8 | GO:0090435 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.3 | 1.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.3 | 0.8 | GO:0021519 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.3 | 0.8 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.3 | 0.8 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.2 | 0.7 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.2 | 2.1 | GO:0009204 | deoxyribonucleoside triphosphate catabolic process(GO:0009204) |
| 0.2 | 1.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.9 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.2 | 3.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.2 | 1.8 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.2 | 1.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 1.5 | GO:2000105 | positive regulation of DNA-dependent DNA replication(GO:2000105) |
| 0.2 | 1.1 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.2 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 1.3 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 1.3 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.2 | 1.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.2 | 0.8 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.2 | 0.6 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.2 | 1.2 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.2 | 0.8 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.2 | 0.6 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.6 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.2 | 1.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 0.5 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.2 | 0.5 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 1.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 1.9 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.2 | 0.5 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.2 | 1.2 | GO:0060420 | box C/D snoRNP assembly(GO:0000492) regulation of heart growth(GO:0060420) |
| 0.2 | 1.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.2 | 3.2 | GO:0044818 | mitotic G2/M transition checkpoint(GO:0044818) |
| 0.2 | 1.1 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.2 | 2.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 0.9 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 | 0.6 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.9 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 1.2 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.7 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 1.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.1 | 1.8 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 1.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.5 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.1 | 1.0 | GO:0002446 | neutrophil mediated immunity(GO:0002446) |
| 0.1 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) positive regulation of DNA binding(GO:0043388) |
| 0.1 | 1.7 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.9 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.1 | 0.6 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.1 | 0.4 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.4 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.1 | 1.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.1 | 1.3 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.6 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.5 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 1.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.2 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.1 | 0.6 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.5 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.9 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.7 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.6 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 0.6 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.1 | 1.6 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 3.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 0.7 | GO:1900118 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 2.5 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.6 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 1.0 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.6 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 2.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.3 | GO:0097240 | telomere localization(GO:0034397) telomere tethering at nuclear periphery(GO:0034398) meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic telomere clustering(GO:0045141) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 1.2 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.5 | GO:0003342 | proepicardium development(GO:0003342) septum transversum development(GO:0003343) determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.1 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.9 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.1 | 1.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.1 | 0.4 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.1 | 0.8 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.8 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 2.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.3 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.1 | 0.7 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.4 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) pronephric proximal tubule development(GO:0035776) |
| 0.1 | 0.6 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.9 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.1 | 2.2 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.1 | 1.4 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.1 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.1 | 1.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.5 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.4 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 2.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 0.6 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.7 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.2 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0060967 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.3 | GO:0072103 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.1 | 0.4 | GO:0043653 | peroxisome fission(GO:0016559) mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.2 | GO:0032263 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 0.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 1.0 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 | 1.1 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.4 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.1 | 0.3 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.1 | 0.9 | GO:0035196 | production of miRNAs involved in gene silencing by miRNA(GO:0035196) |
| 0.1 | 0.5 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.1 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.4 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.1 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.3 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.1 | 0.2 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.1 | 1.1 | GO:0071594 | T cell differentiation in thymus(GO:0033077) thymocyte aggregation(GO:0071594) |
| 0.1 | 5.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.8 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.4 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.1 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.6 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.6 | GO:0010717 | regulation of epithelial to mesenchymal transition(GO:0010717) |
| 0.0 | 0.4 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.0 | 1.8 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.6 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.8 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.5 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 1.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.6 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 2.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.6 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.6 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.7 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 1.4 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.1 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.4 | GO:0050777 | negative regulation of innate immune response(GO:0045824) negative regulation of immune response(GO:0050777) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 1.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 4.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:1901534 | positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 2.2 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.4 | GO:0065005 | plasma lipoprotein particle assembly(GO:0034377) protein-lipid complex assembly(GO:0065005) |
| 0.0 | 0.4 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.3 | GO:0006692 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 1.2 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.3 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 2.2 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0038171 | cannabinoid signaling pathway(GO:0038171) |
| 0.0 | 1.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 1.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0044319 | wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) |
| 0.0 | 0.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 2.3 | GO:0009306 | protein secretion(GO:0009306) |
| 0.0 | 0.2 | GO:0090497 | embryonic medial fin morphogenesis(GO:0035122) mesenchymal cell migration(GO:0090497) |
| 0.0 | 1.4 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.3 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.6 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.0 | 1.6 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0010458 | exit from mitosis(GO:0010458) |
| 0.0 | 1.3 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.7 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.5 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.8 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.3 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.3 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.5 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0070221 | sulfide oxidation(GO:0019418) sulfide oxidation, using sulfide:quinone oxidoreductase(GO:0070221) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 1.0 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.7 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 1.0 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.6 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 1.0 | GO:0045089 | positive regulation of defense response(GO:0031349) positive regulation of innate immune response(GO:0045089) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.8 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 1.3 | GO:0007338 | single fertilization(GO:0007338) |
| 0.0 | 1.0 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 5.6 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 0.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.8 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.4 | GO:0044273 | sulfur compound catabolic process(GO:0044273) |
| 0.0 | 0.0 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 8.7 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 0.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.2 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0030593 | neutrophil chemotaxis(GO:0030593) |
| 0.0 | 0.2 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.5 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 3.1 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 1.1 | GO:0055113 | epiboly involved in gastrulation with mouth forming second(GO:0055113) |
| 0.0 | 1.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 2.5 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.0 | 0.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.5 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.7 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.0 | 1.1 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 0.6 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0061515 | myeloid cell development(GO:0061515) |
| 0.0 | 0.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.8 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.6 | GO:0008380 | RNA splicing(GO:0008380) |
| 0.0 | 0.3 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 2.4 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.3 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.0 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 1.6 | GO:0060026 | convergent extension(GO:0060026) |
| 0.0 | 0.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.9 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.2 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.5 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.1 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.7 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.7 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.0 | 0.2 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell development(GO:0072015) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell development(GO:0072310) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 2.3 | GO:0051301 | cell division(GO:0051301) |
| 0.0 | 0.8 | GO:0030218 | erythrocyte differentiation(GO:0030218) |
| 0.0 | 0.1 | GO:0042493 | response to drug(GO:0042493) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.5 | 1.6 | GO:0033065 | Rad51C-XRCC3 complex(GO:0033065) |
| 0.5 | 2.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.5 | 1.9 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.5 | 2.3 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.4 | 1.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.4 | 2.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.3 | 1.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 1.0 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.3 | 1.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.3 | 0.9 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.3 | 1.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.3 | 5.7 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.3 | 0.8 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.2 | 0.7 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.2 | 0.7 | GO:0031417 | NatC complex(GO:0031417) |
| 0.2 | 1.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 1.5 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.2 | 1.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.2 | 1.3 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.2 | 0.7 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.2 | 0.5 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.2 | 1.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 0.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 0.8 | GO:0034657 | GID complex(GO:0034657) |
| 0.2 | 1.1 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.0 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.1 | 0.6 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 1.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.6 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 1.3 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 4.2 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.3 | GO:0031213 | RSF complex(GO:0031213) |
| 0.1 | 0.4 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 0.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.4 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 1.1 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.7 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 0.9 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.3 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.3 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.8 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.1 | 1.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 0.6 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 0.3 | GO:0098553 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 1.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.5 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.3 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 2.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 1.0 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.4 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.8 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 3.6 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.7 | GO:0030990 | intraciliary transport particle(GO:0030990) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 4.2 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.0 | 3.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 3.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 6.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.2 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.1 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.3 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.4 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 1.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.7 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 5.4 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.5 | 1.6 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.5 | 1.6 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.5 | 1.9 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.5 | 1.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.5 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.4 | 3.6 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.4 | 2.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 4.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.4 | 2.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 0.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.3 | 1.8 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.3 | 1.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.3 | 0.9 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.3 | 1.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.3 | 0.8 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.3 | 1.3 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 2.0 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.2 | 0.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.2 | 0.7 | GO:0072545 | tyrosine binding(GO:0072545) |
| 0.2 | 1.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 1.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 1.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.2 | 1.4 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 0.6 | GO:0042806 | fucose binding(GO:0042806) |
| 0.2 | 0.5 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.2 | 1.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.2 | 0.5 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.2 | 2.8 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.2 | 2.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.5 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.2 | 0.6 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.0 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 0.6 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.2 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.1 | 0.7 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 1.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.5 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.5 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.5 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.1 | 0.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.5 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.9 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 1.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 1.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.1 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.6 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.5 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 1.0 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.1 | 1.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.1 | 1.6 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.4 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) signaling pattern recognition receptor activity(GO:0008329) |
| 0.1 | 0.5 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 0.7 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 2.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.7 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 2.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 1.8 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.4 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.6 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 3 promoter sequence-specific DNA binding(GO:0001006) RNA polymerase III regulatory region DNA binding(GO:0001016) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 1.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 2.3 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.1 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.1 | 1.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.1 | 0.2 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 0.4 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.2 | GO:0016436 | rRNA (uridine) methyltransferase activity(GO:0016436) |
| 0.1 | 0.7 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.8 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 2.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 1.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.1 | 3.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.3 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 0.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.9 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.4 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 1.1 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.3 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 4.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.7 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 3.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 2.1 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 3.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.5 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.9 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 1.1 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.2 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 1.2 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 3.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.4 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.6 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.4 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.4 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.2 | GO:0070137 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.9 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0070224 | sulfide:quinone oxidoreductase activity(GO:0070224) |
| 0.0 | 1.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.5 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.9 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.7 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.3 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.3 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 2.8 | GO:0019783 | ubiquitin-like protein-specific protease activity(GO:0019783) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 2.3 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 7.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 3.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.2 | GO:0015386 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 2.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.3 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.8 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.8 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 11.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.9 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 4.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.0 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 2.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 24.9 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.2 | 2.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 4.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 1.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 1.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 2.1 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.7 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 0.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.9 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 2.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.3 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 2.3 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.1 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 3.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 0.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 0.7 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 2.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 7.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 1.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.6 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.3 | 2.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.2 | 4.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 2.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 1.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 1.8 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.7 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.1 | 1.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 2.0 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 1.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 0.8 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.1 | 0.7 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.1 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.9 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 1.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.7 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.7 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.8 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.1 | 0.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 2.4 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.1 | 2.0 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.1 | 1.0 | REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | Genes involved in Processing of Capped Intronless Pre-mRNA |
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.6 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.8 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.9 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.3 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.4 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME ANTIGEN PROCESSING CROSS PRESENTATION | Genes involved in Antigen processing-Cross presentation |
| 0.0 | 0.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.6 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 2.8 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.5 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.3 | REACTOME SHC MEDIATED CASCADE | Genes involved in SHC-mediated cascade |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.4 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |