Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
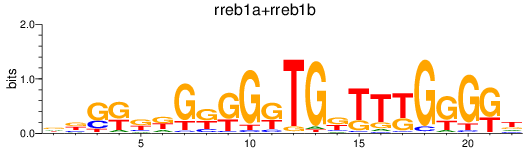
Results for rreb1a+rreb1b
Z-value: 2.44
Transcription factors associated with rreb1a+rreb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rreb1b
|
ENSDARG00000042652 | ras responsive element binding protein 1b |
|
rreb1a
|
ENSDARG00000063701 | ras responsive element binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rreb1a | dr11_v1_chr24_-_2450597_2450597 | 0.39 | 1.0e-01 | Click! |
| rreb1b | dr11_v1_chr2_+_21048661_21048661 | 0.32 | 1.9e-01 | Click! |
Activity profile of rreb1a+rreb1b motif
Sorted Z-values of rreb1a+rreb1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_828663 | 11.25 |
ENSDART00000181776
|
BX537263.4
|
|
| chr5_+_817016 | 11.25 |
ENSDART00000188498
|
BX537263.3
|
|
| chr13_-_638485 | 6.54 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr2_-_22535 | 6.24 |
ENSDART00000157877
|
CABZ01092282.1
|
|
| chr7_-_10606 | 4.26 |
ENSDART00000192650
ENSDART00000186761 |
FO704772.2
|
|
| chr9_+_51882534 | 4.09 |
ENSDART00000165493
|
CABZ01079550.1
|
|
| chr4_+_90048 | 3.98 |
ENSDART00000166440
|
lrp6
|
low density lipoprotein receptor-related protein 6 |
| chr20_+_52595220 | 3.62 |
ENSDART00000180610
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr24_-_38110779 | 3.61 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr7_-_57949117 | 3.52 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr4_+_77735212 | 3.47 |
ENSDART00000160716
|
si:dkey-238k10.1
|
si:dkey-238k10.1 |
| chr11_-_4235811 | 3.38 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr19_+_56351 | 3.30 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr13_-_11536951 | 3.13 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr20_-_1847433 | 2.87 |
ENSDART00000164211
|
CABZ01065425.1
|
|
| chr10_+_16756546 | 2.75 |
ENSDART00000168624
|
CABZ01024659.1
|
|
| chr1_-_8012476 | 2.60 |
ENSDART00000177051
|
CR855320.3
|
|
| chr20_-_25518488 | 2.60 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr12_+_13244149 | 2.44 |
ENSDART00000186984
ENSDART00000105896 |
atp2a1l
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1, like |
| chr19_+_1510971 | 2.39 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr8_+_554531 | 2.36 |
ENSDART00000193623
|
FO704758.2
|
|
| chr12_+_47162761 | 2.35 |
ENSDART00000192339
ENSDART00000167726 |
RYR2
|
ryanodine receptor 2 |
| chr7_+_39634873 | 2.30 |
ENSDART00000114774
|
ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr17_-_20711735 | 2.27 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr7_+_7630409 | 2.27 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr2_+_7192966 | 2.25 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr15_-_47193564 | 2.22 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr19_+_43256986 | 2.21 |
ENSDART00000182336
|
DGKD
|
diacylglycerol kinase delta |
| chr12_+_47162456 | 2.18 |
ENSDART00000186272
ENSDART00000180811 |
RYR2
|
ryanodine receptor 2 |
| chr15_-_42736433 | 2.10 |
ENSDART00000154379
|
si:ch211-181d7.1
|
si:ch211-181d7.1 |
| chr21_-_45613564 | 2.08 |
ENSDART00000160324
|
LO018363.1
|
|
| chr4_-_53370 | 2.06 |
ENSDART00000180254
ENSDART00000186529 |
CU856344.2
|
|
| chr6_+_54239625 | 2.04 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr23_+_3591690 | 2.00 |
ENSDART00000180822
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr16_+_53387085 | 1.93 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr2_+_22694382 | 1.90 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr23_+_3616224 | 1.90 |
ENSDART00000190917
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr7_+_38717624 | 1.88 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr13_+_51853716 | 1.88 |
ENSDART00000175341
ENSDART00000187855 |
LT631684.1
|
|
| chr23_+_3587230 | 1.85 |
ENSDART00000055103
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr23_-_38054 | 1.85 |
ENSDART00000170393
|
CABZ01074076.1
|
|
| chr5_-_201600 | 1.80 |
ENSDART00000158495
|
CABZ01088906.1
|
|
| chr23_+_22873415 | 1.79 |
ENSDART00000135130
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr21_+_45510448 | 1.77 |
ENSDART00000160494
ENSDART00000167914 |
fnip1
|
folliculin interacting protein 1 |
| chr17_+_12075805 | 1.76 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr20_+_18943406 | 1.76 |
ENSDART00000193590
|
mtmr9
|
myotubularin related protein 9 |
| chr11_-_44037692 | 1.74 |
ENSDART00000186333
|
FP016005.2
|
|
| chr22_+_1186866 | 1.73 |
ENSDART00000167710
|
eps8l3a
|
EPS8-like 3a |
| chr18_+_9637744 | 1.68 |
ENSDART00000190171
|
pclob
|
piccolo presynaptic cytomatrix protein b |
| chr6_+_58492201 | 1.68 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr20_+_4392687 | 1.67 |
ENSDART00000187271
|
im:7142702
|
im:7142702 |
| chr9_+_36946340 | 1.66 |
ENSDART00000135281
|
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr13_+_12801092 | 1.65 |
ENSDART00000164040
ENSDART00000134725 |
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr14_-_1565317 | 1.63 |
ENSDART00000169496
|
CABZ01064248.1
|
|
| chr8_+_54081819 | 1.59 |
ENSDART00000005857
ENSDART00000161795 |
prickle2a
|
prickle homolog 2a |
| chr2_-_50298337 | 1.59 |
ENSDART00000155125
|
cntnap2b
|
contactin associated protein like 2b |
| chr19_+_2279051 | 1.57 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr22_+_438714 | 1.53 |
ENSDART00000136491
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr3_+_60007703 | 1.53 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr16_-_13662514 | 1.51 |
ENSDART00000146348
|
shisa7a
|
shisa family member 7a |
| chr21_+_28445052 | 1.50 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr17_+_691453 | 1.49 |
ENSDART00000159271
|
fancm
|
Fanconi anemia, complementation group M |
| chr4_-_1720648 | 1.48 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr5_+_19337108 | 1.48 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr7_-_55454406 | 1.47 |
ENSDART00000108646
|
piezo1
|
piezo-type mechanosensitive ion channel component 1 |
| chr23_-_15284757 | 1.40 |
ENSDART00000139135
|
sulf2b
|
sulfatase 2b |
| chr13_+_1182257 | 1.40 |
ENSDART00000033528
ENSDART00000183702 ENSDART00000147959 |
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr21_+_170038 | 1.40 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr10_-_74408 | 1.38 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr22_+_413349 | 1.38 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr5_+_71802014 | 1.38 |
ENSDART00000124939
ENSDART00000097164 |
LHX3
|
LIM homeobox 3 |
| chr15_+_46108548 | 1.37 |
ENSDART00000099023
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr25_+_12012 | 1.37 |
ENSDART00000170348
|
cntn1a
|
contactin 1a |
| chr22_-_18112374 | 1.37 |
ENSDART00000191154
|
ncanb
|
neurocan b |
| chr8_-_5847533 | 1.37 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr12_+_48480632 | 1.37 |
ENSDART00000158157
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr10_+_23537576 | 1.36 |
ENSDART00000130515
|
CR847844.1
|
|
| chr20_-_54462551 | 1.34 |
ENSDART00000171769
ENSDART00000169692 |
evlb
|
Enah/Vasp-like b |
| chr11_+_45461419 | 1.32 |
ENSDART00000173424
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr22_+_997838 | 1.31 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr23_-_4855122 | 1.30 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr10_+_2234283 | 1.30 |
ENSDART00000136363
|
cntnap3
|
contactin associated protein like 3 |
| chr15_-_28618502 | 1.27 |
ENSDART00000086902
|
slc6a4a
|
solute carrier family 6 (neurotransmitter transporter), member 4a |
| chr16_-_52821023 | 1.26 |
ENSDART00000074718
|
spire1b
|
spire-type actin nucleation factor 1b |
| chr11_+_38280454 | 1.26 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr23_-_969844 | 1.25 |
ENSDART00000127037
|
cdh26.2
|
cadherin 26, tandem duplicate 2 |
| chr10_+_45302425 | 1.25 |
ENSDART00000159954
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr18_+_22793743 | 1.19 |
ENSDART00000150106
|
gnao1a
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a |
| chr4_+_9279515 | 1.19 |
ENSDART00000048707
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr2_-_51700709 | 1.18 |
ENSDART00000188601
|
tgm1l1
|
transglutaminase 1 like 1 |
| chr21_+_6212844 | 1.18 |
ENSDART00000150301
|
fnbp1b
|
formin binding protein 1b |
| chr18_-_50845804 | 1.17 |
ENSDART00000158517
|
PDPR
|
si:cabz01113374.3 |
| chr15_-_47479119 | 1.16 |
ENSDART00000164957
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr25_-_1124851 | 1.16 |
ENSDART00000067558
|
spg11
|
spastic paraplegia 11 |
| chr4_-_73162005 | 1.15 |
ENSDART00000164127
|
si:ch73-170d6.4
|
si:ch73-170d6.4 |
| chr1_+_47585700 | 1.13 |
ENSDART00000153746
ENSDART00000084457 |
sh3pxd2aa
|
SH3 and PX domains 2Aa |
| chr8_-_1698155 | 1.13 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr18_+_14342326 | 1.13 |
ENSDART00000181013
ENSDART00000138372 |
si:dkey-246g23.2
|
si:dkey-246g23.2 |
| chr15_-_434503 | 1.13 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr19_+_233143 | 1.12 |
ENSDART00000175273
|
syngap1a
|
synaptic Ras GTPase activating protein 1a |
| chr9_-_12493628 | 1.12 |
ENSDART00000180832
|
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr20_-_4407228 | 1.12 |
ENSDART00000166997
|
CABZ01032982.1
|
|
| chr18_-_50083405 | 1.11 |
ENSDART00000098668
ENSDART00000182215 ENSDART00000165423 |
abcc8b
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8b |
| chr5_+_23630384 | 1.11 |
ENSDART00000013745
|
cx39.9
|
connexin 39.9 |
| chr20_-_54192130 | 1.11 |
ENSDART00000020834
|
CABZ01111496.1
|
|
| chr8_+_54284961 | 1.10 |
ENSDART00000122692
|
plxnd1
|
plexin D1 |
| chr21_+_3854414 | 1.10 |
ENSDART00000122699
|
miga2
|
mitoguardin 2 |
| chr6_+_2271559 | 1.10 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr14_-_1432875 | 1.09 |
ENSDART00000164408
ENSDART00000159774 |
zgc:152774
|
zgc:152774 |
| chr3_-_62380146 | 1.09 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr21_-_45728069 | 1.08 |
ENSDART00000185101
|
CU302316.1
|
|
| chr2_-_44183613 | 1.08 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr7_-_73752955 | 1.08 |
ENSDART00000171254
ENSDART00000009888 |
casq1b
|
calsequestrin 1b |
| chr20_-_16972351 | 1.08 |
ENSDART00000148312
ENSDART00000186702 |
si:ch73-74h11.1
|
si:ch73-74h11.1 |
| chr2_-_5776030 | 1.07 |
ENSDART00000133851
|
nyap2b
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2b |
| chr4_+_20263097 | 1.07 |
ENSDART00000138820
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr3_-_1434135 | 1.06 |
ENSDART00000149622
|
mgp
|
matrix Gla protein |
| chr25_+_37268900 | 1.06 |
ENSDART00000156737
|
si:dkey-234i14.6
|
si:dkey-234i14.6 |
| chr22_+_39074688 | 1.06 |
ENSDART00000153547
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr2_+_48073972 | 1.05 |
ENSDART00000186442
|
klf6b
|
Kruppel-like factor 6b |
| chr25_-_37465064 | 1.05 |
ENSDART00000186128
|
zgc:158366
|
zgc:158366 |
| chr16_+_41826584 | 1.05 |
ENSDART00000147523
|
si:dkey-199f5.6
|
si:dkey-199f5.6 |
| chr12_+_33396489 | 1.04 |
ENSDART00000149960
|
fasn
|
fatty acid synthase |
| chr6_-_43677125 | 1.04 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr6_-_60031693 | 1.04 |
ENSDART00000160275
|
CABZ01079262.1
|
|
| chr22_-_38459316 | 1.03 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr10_+_213878 | 1.03 |
ENSDART00000135903
ENSDART00000138812 |
mpzl1l
|
myelin protein zero-like 1 like |
| chr3_-_5664123 | 1.03 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr5_-_26181863 | 1.02 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_+_21891305 | 1.02 |
ENSDART00000136788
|
si:ch73-92i20.1
|
si:ch73-92i20.1 |
| chr1_+_176583 | 1.02 |
ENSDART00000168760
ENSDART00000160425 |
LAMP1
|
lysosomal associated membrane protein 1 |
| chr11_+_37049347 | 1.00 |
ENSDART00000109235
|
bicd2
|
bicaudal D homolog 2 (Drosophila) |
| chr18_-_49078428 | 0.99 |
ENSDART00000160702
ENSDART00000174103 |
BX663503.3
|
|
| chr23_-_478201 | 0.99 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr12_+_42436328 | 0.99 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr10_+_428269 | 0.99 |
ENSDART00000140715
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr9_+_4306122 | 0.98 |
ENSDART00000193722
ENSDART00000190521 |
kalrna
|
kalirin RhoGEF kinase a |
| chr17_+_5931530 | 0.98 |
ENSDART00000168326
ENSDART00000189790 |
znf513b
|
zinc finger protein 513b |
| chr11_+_37803773 | 0.98 |
ENSDART00000019047
|
sox13
|
SRY (sex determining region Y)-box 13 |
| chr6_+_59967994 | 0.98 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr10_-_641609 | 0.98 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr5_-_3839285 | 0.97 |
ENSDART00000122292
|
mlxipl
|
MLX interacting protein like |
| chr23_-_306796 | 0.97 |
ENSDART00000143125
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr8_-_1909840 | 0.96 |
ENSDART00000147408
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr16_-_35329803 | 0.95 |
ENSDART00000161729
ENSDART00000157700 ENSDART00000184584 ENSDART00000174713 ENSDART00000162518 |
ptprub
|
protein tyrosine phosphatase, receptor type, U, b |
| chr19_-_26943936 | 0.95 |
ENSDART00000159319
|
zbtb12.1
|
zinc finger and BTB domain containing 12, tandem duplicate 1 |
| chr16_+_22587661 | 0.94 |
ENSDART00000129612
ENSDART00000142241 |
she
|
Src homology 2 domain containing E |
| chr8_-_22639794 | 0.93 |
ENSDART00000188029
|
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr2_-_44183451 | 0.91 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr24_-_21470766 | 0.90 |
ENSDART00000178121
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr12_+_23892972 | 0.90 |
ENSDART00000152852
|
svila
|
supervillin a |
| chr6_-_58901552 | 0.90 |
ENSDART00000122003
|
mbd6
|
methyl-CpG binding domain protein 6 |
| chr14_+_26759332 | 0.89 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr15_-_26568278 | 0.89 |
ENSDART00000182609
|
wdr81
|
WD repeat domain 81 |
| chr20_-_1635922 | 0.88 |
ENSDART00000181502
|
CR846082.1
|
|
| chr10_+_1849874 | 0.88 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr15_-_40175894 | 0.88 |
ENSDART00000156632
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr5_+_17780475 | 0.87 |
ENSDART00000110783
ENSDART00000115227 |
chfr
|
checkpoint with forkhead and ring finger domains, E3 ubiquitin protein ligase |
| chr3_+_1107102 | 0.86 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr23_-_21758253 | 0.86 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr9_+_32428679 | 0.86 |
ENSDART00000187650
ENSDART00000190472 |
plcl1
|
phospholipase C like 1 |
| chr25_-_37501371 | 0.86 |
ENSDART00000160498
|
CABZ01088346.1
|
|
| chr7_+_1306723 | 0.85 |
ENSDART00000189868
|
si:dkeyp-74b6.2
|
si:dkeyp-74b6.2 |
| chr23_-_1348933 | 0.85 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr7_-_54430505 | 0.84 |
ENSDART00000167905
|
ano1
|
anoctamin 1, calcium activated chloride channel |
| chr10_+_3875716 | 0.84 |
ENSDART00000189268
ENSDART00000180624 |
TTC28
|
tetratricopeptide repeat domain 28 |
| chr5_-_28029558 | 0.83 |
ENSDART00000078649
|
abcb9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr8_+_37489495 | 0.82 |
ENSDART00000141516
|
fmodb
|
fibromodulin b |
| chr16_+_10918252 | 0.82 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr19_-_5380770 | 0.81 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr7_-_5191797 | 0.81 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr18_+_40572294 | 0.80 |
ENSDART00000147003
|
si:ch211-132b12.2
|
si:ch211-132b12.2 |
| chr2_-_41620112 | 0.80 |
ENSDART00000138822
ENSDART00000004816 |
zgc:110158
|
zgc:110158 |
| chr18_-_50579629 | 0.80 |
ENSDART00000188252
|
zgc:158464
|
zgc:158464 |
| chr23_-_44233408 | 0.79 |
ENSDART00000149318
ENSDART00000085528 |
zgc:158659
|
zgc:158659 |
| chr9_-_23807032 | 0.79 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr5_+_32009080 | 0.78 |
ENSDART00000186885
|
scai
|
suppressor of cancer cell invasion |
| chr23_+_2914577 | 0.78 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr4_+_9279784 | 0.77 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr14_-_41556533 | 0.76 |
ENSDART00000074401
|
itga6l
|
integrin, alpha 6, like |
| chr2_+_52847049 | 0.75 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr14_-_853176 | 0.75 |
ENSDART00000041988
|
slc34a1a
|
solute carrier family 34 (type II sodium/phosphate cotransporter), member 1a |
| chr5_+_480119 | 0.75 |
ENSDART00000055681
|
tek
|
TEK tyrosine kinase, endothelial |
| chr9_-_52609336 | 0.75 |
ENSDART00000114805
|
tmem169b
|
transmembrane protein 169b |
| chr1_+_109798 | 0.75 |
ENSDART00000165402
|
F7 (1 of many)
|
zgc:163025 |
| chr15_+_47362728 | 0.74 |
ENSDART00000180712
|
CABZ01099833.1
|
|
| chr6_-_442163 | 0.74 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr5_-_30080332 | 0.73 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr16_+_68069 | 0.73 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr5_-_417495 | 0.72 |
ENSDART00000180586
ENSDART00000189408 |
HOOK3
|
hook microtubule tethering protein 3 |
| chr11_+_23760470 | 0.72 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr2_-_59376735 | 0.71 |
ENSDART00000193624
|
ftr38
|
finTRIM family, member 38 |
| chr2_+_33368414 | 0.71 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr3_-_38692920 | 0.70 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr9_+_36314867 | 0.70 |
ENSDART00000176763
|
lrp1bb
|
low density lipoprotein receptor-related protein 1Bb |
| chr8_+_1715 | 0.69 |
ENSDART00000181131
|
tmed7
|
transmembrane p24 trafficking protein 7 |
| chr13_-_20519001 | 0.69 |
ENSDART00000168955
|
gfra1a
|
gdnf family receptor alpha 1a |
| chr20_-_1560724 | 0.69 |
ENSDART00000179193
ENSDART00000092508 ENSDART00000133369 |
ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr19_+_42754882 | 0.69 |
ENSDART00000133711
|
clasp2
|
cytoplasmic linker associated protein 2 |
| chr5_+_337215 | 0.69 |
ENSDART00000167982
|
rnf170
|
ring finger protein 170 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rreb1a+rreb1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.6 | 1.8 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.5 | 1.5 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.4 | 1.5 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.4 | 1.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.3 | 1.0 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.3 | 1.6 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.3 | 1.3 | GO:0051610 | serotonin transport(GO:0006837) serotonin uptake(GO:0051610) |
| 0.3 | 0.9 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.3 | 1.5 | GO:0010482 | ventriculo bulbo valve development(GO:0003173) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.3 | 1.7 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.3 | 6.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 1.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 3.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.2 | 0.7 | GO:0009750 | response to fructose(GO:0009750) |
| 0.2 | 1.8 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.2 | 0.9 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.2 | 1.5 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.2 | 2.1 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.2 | 0.6 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.2 | 1.9 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.2 | 0.6 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.6 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.2 | 1.3 | GO:0033206 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.7 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.2 | 1.7 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.2 | 1.5 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.2 | 1.7 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.2 | 4.2 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) |
| 0.1 | 1.5 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 0.4 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 1.0 | GO:2000095 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.8 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.6 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.2 | GO:0061217 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 2.0 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.1 | 0.4 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.1 | 1.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.3 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.1 | 0.9 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 1.4 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.5 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.9 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.7 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 1.0 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.5 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.8 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.8 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.3 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 0.4 | GO:0098957 | anterograde axonal transport of mitochondrion(GO:0098957) |
| 0.1 | 1.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.5 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.1 | 1.4 | GO:0050796 | regulation of insulin secretion(GO:0050796) |
| 0.1 | 0.2 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 2.0 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.5 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 2.4 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.1 | 7.0 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 1.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.1 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 2.7 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.4 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.3 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 0.5 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 1.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.0 | 2.6 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 1.1 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 1.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.4 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.0 | 1.1 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.6 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 1.2 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.7 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.0 | 0.8 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.4 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.0 | 0.1 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 1.7 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 2.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.7 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 1.6 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.4 | GO:0040036 | regulation of fibroblast growth factor receptor signaling pathway(GO:0040036) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 2.1 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.6 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.0 | GO:0060039 | pericardium development(GO:0060039) |
| 0.0 | 2.8 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.4 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.1 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.5 | GO:0009880 | embryonic pattern specification(GO:0009880) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 1.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.2 | 0.6 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.2 | 1.1 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 0.9 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.7 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.2 | 0.8 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.2 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.9 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 2.7 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 4.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 1.1 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 3.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 3.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 1.5 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 4.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.9 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.8 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.7 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 2.8 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.9 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 4.1 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 1.8 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.4 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.8 | GO:0071253 | connexin binding(GO:0071253) |
| 0.4 | 1.3 | GO:0015222 | serotonin:sodium symporter activity(GO:0005335) serotonin transmembrane transporter activity(GO:0015222) |
| 0.4 | 1.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.4 | 1.8 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.3 | 1.5 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.8 | GO:0046978 | TAP1 binding(GO:0046978) |
| 0.3 | 4.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.2 | 1.2 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.2 | 1.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.2 | 3.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.2 | 0.6 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.2 | 3.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.2 | 1.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 2.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.2 | 2.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 1.1 | GO:0016176 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 0.6 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 1.7 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.1 | 0.5 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.7 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.1 | 1.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.7 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.8 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 3.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 1.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 1.0 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.8 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.6 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.1 | 1.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.5 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.7 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 2.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 2.7 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.8 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 1.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 2.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.6 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.5 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 2.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.2 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 1.1 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 4.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.6 | GO:0004601 | peroxidase activity(GO:0004601) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.9 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 1.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.4 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 3.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 0.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 0.9 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.7 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.7 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.4 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.4 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 3.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 1.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 1.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 1.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 0.9 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.4 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 1.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |