Project
PRJEB1986: zebrafish developmental stages transcriptome
Navigation
Downloads
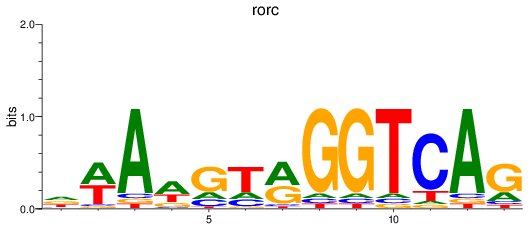
Results for rorc
Z-value: 0.93
Transcription factors associated with rorc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rorc
|
ENSDARG00000087195 | RAR-related orphan receptor C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rorc | dr11_v1_chr16_+_17913936_17913945 | -0.35 | 1.4e-01 | Click! |
Activity profile of rorc motif
Sorted Z-values of rorc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_43314700 | 5.17 |
ENSDART00000049045
ENSDART00000133158 |
matn1
|
matrilin 1 |
| chr14_+_17376940 | 2.90 |
ENSDART00000054590
ENSDART00000010148 |
spon2b
|
spondin 2b, extracellular matrix protein |
| chr15_+_19682013 | 2.62 |
ENSDART00000127368
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr6_+_13742899 | 2.43 |
ENSDART00000104722
|
cdk5r2a
|
cyclin-dependent kinase 5, regulatory subunit 2a (p39) |
| chr15_+_19681718 | 1.85 |
ENSDART00000164803
|
si:dkey-4p15.5
|
si:dkey-4p15.5 |
| chr20_+_16750177 | 1.66 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr15_-_37829160 | 1.64 |
ENSDART00000099425
|
ctrl
|
chymotrypsin-like |
| chr5_+_2815021 | 1.63 |
ENSDART00000020472
|
hpda
|
4-hydroxyphenylpyruvate dioxygenase a |
| chr19_+_4990320 | 1.59 |
ENSDART00000147056
|
zgc:91968
|
zgc:91968 |
| chr21_+_27189490 | 1.58 |
ENSDART00000125349
|
bada
|
BCL2 associated agonist of cell death a |
| chr19_+_4990496 | 1.53 |
ENSDART00000151050
ENSDART00000017535 |
zgc:91968
|
zgc:91968 |
| chr8_+_25351863 | 1.52 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr10_-_39321367 | 1.40 |
ENSDART00000129647
|
smtlb
|
somatolactin beta |
| chr23_+_19564392 | 1.33 |
ENSDART00000144746
|
atp6ap1lb
|
ATPase H+ transporting accessory protein 1 like b |
| chr5_-_11573490 | 1.23 |
ENSDART00000109577
|
FO704871.1
|
|
| chr21_+_40106448 | 1.22 |
ENSDART00000100166
|
serpinf1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr17_-_6738538 | 1.14 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr13_+_29470442 | 1.13 |
ENSDART00000028417
|
lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 |
| chr4_+_17280868 | 1.11 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr20_-_14680897 | 1.03 |
ENSDART00000063857
ENSDART00000161314 |
scrn2
|
secernin 2 |
| chr24_-_32408404 | 1.03 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr11_-_24458786 | 1.01 |
ENSDART00000089713
|
mxra8a
|
matrix-remodelling associated 8a |
| chr11_+_30672282 | 1.01 |
ENSDART00000023981
ENSDART00000188846 |
ttbk1a
|
tau tubulin kinase 1a |
| chr7_+_65240227 | 0.96 |
ENSDART00000168287
|
bco1l
|
beta-carotene oxygenase 1, like |
| chr23_-_36724575 | 0.92 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr19_+_30990129 | 0.90 |
ENSDART00000052169
ENSDART00000193376 |
sync
|
syncoilin, intermediate filament protein |
| chr19_+_30990815 | 0.87 |
ENSDART00000134645
|
sync
|
syncoilin, intermediate filament protein |
| chr22_-_13544244 | 0.86 |
ENSDART00000110136
|
cntnap5b
|
contactin associated protein-like 5b |
| chr3_+_1016404 | 0.81 |
ENSDART00000189545
|
si:ch1073-464p5.5
|
si:ch1073-464p5.5 |
| chr22_-_38480186 | 0.79 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr8_-_29706882 | 0.79 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr21_+_30549512 | 0.78 |
ENSDART00000132831
|
rab38c
|
RAB38c, member of RAS oncogene family |
| chr5_-_16983336 | 0.78 |
ENSDART00000038740
|
galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9 |
| chr4_-_4129187 | 0.78 |
ENSDART00000146889
|
asb15b
|
ankyrin repeat and SOCS box containing 15b |
| chr2_+_28889936 | 0.73 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr9_+_35876927 | 0.72 |
ENSDART00000138834
|
mab21l3
|
mab-21-like 3 |
| chr2_+_49417900 | 0.71 |
ENSDART00000122742
ENSDART00000160783 |
rorcb
|
RAR-related orphan receptor C b |
| chr12_+_1286642 | 0.68 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr24_+_24809877 | 0.68 |
ENSDART00000185117
|
dnajc5b
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
| chr22_-_600016 | 0.63 |
ENSDART00000086434
|
tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr15_-_12360409 | 0.63 |
ENSDART00000164596
|
tmprss13a
|
transmembrane protease, serine 13a |
| chr5_-_30079434 | 0.62 |
ENSDART00000133981
|
bco2a
|
beta-carotene oxygenase 2a |
| chr18_+_7594012 | 0.60 |
ENSDART00000062150
|
zgc:77752
|
zgc:77752 |
| chr22_-_11124419 | 0.59 |
ENSDART00000149634
|
atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr23_+_44741500 | 0.58 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr15_-_24178893 | 0.56 |
ENSDART00000077980
|
pipox
|
pipecolic acid oxidase |
| chr1_+_9708801 | 0.55 |
ENSDART00000189621
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr2_-_56655769 | 0.53 |
ENSDART00000113589
|
gpx4b
|
glutathione peroxidase 4b |
| chr16_+_26738404 | 0.53 |
ENSDART00000163110
|
fsbp
|
fibrinogen silencer binding protein |
| chr13_+_21768447 | 0.52 |
ENSDART00000100941
|
chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr25_-_32888115 | 0.52 |
ENSDART00000087586
|
c2cd4a
|
C2 calcium dependent domain containing 4A |
| chr19_-_8096984 | 0.51 |
ENSDART00000146987
|
si:dkey-266f7.9
|
si:dkey-266f7.9 |
| chr18_+_22302635 | 0.51 |
ENSDART00000141051
|
carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr12_+_16106459 | 0.50 |
ENSDART00000114174
|
muc13b
|
mucin 13b, cell surface associated |
| chr25_-_17918810 | 0.50 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr10_+_2232023 | 0.49 |
ENSDART00000097695
|
cntnap3
|
contactin associated protein like 3 |
| chr11_+_28476298 | 0.49 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr21_-_40557281 | 0.47 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr7_+_66048102 | 0.47 |
ENSDART00000104523
|
arntl1b
|
aryl hydrocarbon receptor nuclear translocator-like 1b |
| chr3_-_22829710 | 0.45 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr18_-_42313798 | 0.45 |
ENSDART00000098639
|
cntn5
|
contactin 5 |
| chr21_-_5881344 | 0.44 |
ENSDART00000009241
|
rpl35
|
ribosomal protein L35 |
| chr17_-_2126517 | 0.44 |
ENSDART00000013093
|
zfyve19
|
zinc finger, FYVE domain containing 19 |
| chr3_-_26191960 | 0.44 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr7_-_28058442 | 0.43 |
ENSDART00000173842
|
si:ch211-235p24.2
|
si:ch211-235p24.2 |
| chr7_+_24573721 | 0.42 |
ENSDART00000173938
ENSDART00000173681 |
si:dkeyp-75h12.7
|
si:dkeyp-75h12.7 |
| chr15_+_42599501 | 0.42 |
ENSDART00000177646
|
grik1b
|
glutamate receptor, ionotropic, kainate 1b |
| chr9_+_21993092 | 0.41 |
ENSDART00000059693
|
crygm7
|
crystallin, gamma M7 |
| chr8_+_25352268 | 0.41 |
ENSDART00000187829
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr15_-_31419805 | 0.41 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr24_-_31422574 | 0.39 |
ENSDART00000158567
|
cngb3.1
|
cyclic nucleotide gated channel beta 3, tandem duplicate 1 |
| chr19_-_8604429 | 0.38 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr16_+_30438041 | 0.38 |
ENSDART00000137977
|
pear1
|
platelet endothelial aggregation receptor 1 |
| chr11_-_29737088 | 0.37 |
ENSDART00000159828
|
si:ch211-218g23.6
|
si:ch211-218g23.6 |
| chr24_-_35707552 | 0.36 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr2_-_56649883 | 0.35 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr16_-_24561354 | 0.35 |
ENSDART00000193278
ENSDART00000126274 |
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr17_-_13072334 | 0.34 |
ENSDART00000159598
|
CU469462.1
|
|
| chr25_-_17918536 | 0.34 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr21_+_41649336 | 0.32 |
ENSDART00000164694
ENSDART00000181539 |
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr12_-_32013125 | 0.32 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr9_-_48937089 | 0.32 |
ENSDART00000193442
|
cers6
|
ceramide synthase 6 |
| chr2_+_48282590 | 0.29 |
ENSDART00000035338
|
lpar5a
|
lysophosphatidic acid receptor 5a |
| chr3_-_5362347 | 0.29 |
ENSDART00000187511
|
trim35-7
|
tripartite motif containing 35-7 |
| chr22_-_5752009 | 0.28 |
ENSDART00000190052
|
bcdin3d
|
BCDIN3 domain containing |
| chr9_+_13682133 | 0.27 |
ENSDART00000175639
|
mpp4a
|
membrane protein, palmitoylated 4a (MAGUK p55 subfamily member 4) |
| chr22_+_8612462 | 0.27 |
ENSDART00000114586
|
CR450686.1
|
|
| chr1_-_33353778 | 0.26 |
ENSDART00000041191
|
gyg2
|
glycogenin 2 |
| chr6_-_39605734 | 0.25 |
ENSDART00000044276
ENSDART00000179059 |
dip2bb
|
disco-interacting protein 2 homolog Bb |
| chr3_-_31226841 | 0.25 |
ENSDART00000085903
|
syt17
|
synaptotagmin XVII |
| chr19_+_2590182 | 0.25 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr17_-_37312922 | 0.24 |
ENSDART00000153555
|
si:ch211-180n24.3
|
si:ch211-180n24.3 |
| chr11_-_1948784 | 0.24 |
ENSDART00000082475
|
nr1d4b
|
nuclear receptor subfamily 1, group D, member 4b |
| chr8_-_51349429 | 0.23 |
ENSDART00000134043
ENSDART00000098282 |
si:ch211-198o12.4
|
si:ch211-198o12.4 |
| chr3_-_39198113 | 0.23 |
ENSDART00000102690
|
RETSAT
|
zgc:154169 |
| chr17_-_27976031 | 0.22 |
ENSDART00000154829
|
ptafr
|
platelet-activating factor receptor |
| chr17_-_12718547 | 0.22 |
ENSDART00000193002
|
BX294375.1
|
|
| chr7_-_22632518 | 0.21 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr21_-_25613249 | 0.21 |
ENSDART00000137896
|
fibpb
|
fibroblast growth factor (acidic) intracellular binding protein b |
| chr22_+_26263290 | 0.21 |
ENSDART00000184840
|
BX649315.1
|
|
| chr8_+_21146262 | 0.20 |
ENSDART00000045684
|
porcn
|
porcupine O-acyltransferase |
| chr6_-_8244474 | 0.20 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr25_-_19374710 | 0.20 |
ENSDART00000184483
ENSDART00000188706 |
map1ab
|
microtubule-associated protein 1Ab |
| chr23_-_5683147 | 0.19 |
ENSDART00000102766
ENSDART00000067351 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr22_+_36704764 | 0.18 |
ENSDART00000146662
|
trim35-33
|
tripartite motif containing 35-33 |
| chr5_-_31689796 | 0.18 |
ENSDART00000184319
ENSDART00000190229 ENSDART00000186294 ENSDART00000170313 ENSDART00000147065 ENSDART00000134427 ENSDART00000098172 |
sh3glb2b
|
SH3-domain GRB2-like endophilin B2b |
| chr19_+_41173386 | 0.18 |
ENSDART00000142773
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr6_-_43005901 | 0.18 |
ENSDART00000161071
|
si:ch73-361p23.3
|
si:ch73-361p23.3 |
| chr24_-_982443 | 0.15 |
ENSDART00000063151
|
napga
|
N-ethylmaleimide-sensitive factor attachment protein, gamma a |
| chr8_+_18464235 | 0.14 |
ENSDART00000110571
|
alkal1
|
ALK and LTK ligand 1 |
| chr25_+_30233638 | 0.14 |
ENSDART00000192558
ENSDART00000067057 ENSDART00000147327 ENSDART00000145924 ENSDART00000148250 |
alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr20_-_4738101 | 0.14 |
ENSDART00000050201
ENSDART00000152559 ENSDART00000053858 ENSDART00000125620 |
papola
|
poly(A) polymerase alpha |
| chr5_-_39698224 | 0.13 |
ENSDART00000076929
|
prkg2
|
protein kinase, cGMP-dependent, type II |
| chr25_+_3035384 | 0.13 |
ENSDART00000184219
ENSDART00000149360 |
mpi
|
mannose phosphate isomerase |
| chr2_-_25120197 | 0.13 |
ENSDART00000132549
|
pccb
|
propionyl CoA carboxylase, beta polypeptide |
| chr7_-_22632938 | 0.13 |
ENSDART00000159867
ENSDART00000165706 |
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr2_-_8648440 | 0.13 |
ENSDART00000135743
|
si:ch211-71m22.3
|
si:ch211-71m22.3 |
| chr11_+_26363435 | 0.12 |
ENSDART00000088800
|
ppcs
|
phosphopantothenoylcysteine synthetase |
| chr21_-_20725853 | 0.12 |
ENSDART00000114502
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr17_+_2214283 | 0.12 |
ENSDART00000189187
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr20_+_44082532 | 0.12 |
ENSDART00000188481
|
BX510352.1
|
|
| chr1_+_19433004 | 0.11 |
ENSDART00000133959
|
clockb
|
clock circadian regulator b |
| chr15_-_34322915 | 0.11 |
ENSDART00000190543
ENSDART00000019651 ENSDART00000193601 |
dgkb
|
diacylglycerol kinase, beta |
| chr11_+_19159083 | 0.11 |
ENSDART00000138964
|
adamts9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
| chr20_-_30377221 | 0.11 |
ENSDART00000126229
|
rps7
|
ribosomal protein S7 |
| chr15_-_34878388 | 0.11 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr5_+_70155935 | 0.11 |
ENSDART00000165570
|
rgs3a
|
regulator of G protein signaling 3a |
| chr8_+_18463797 | 0.10 |
ENSDART00000148958
|
alkal1
|
ALK and LTK ligand 1 |
| chr12_+_31616412 | 0.10 |
ENSDART00000124439
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr15_-_2841677 | 0.08 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr3_-_5555500 | 0.08 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr6_-_49159207 | 0.07 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr3_+_32698424 | 0.07 |
ENSDART00000055340
|
fus
|
FUS RNA binding protein |
| chr25_-_1235457 | 0.07 |
ENSDART00000093093
|
coro2bb
|
coronin, actin binding protein, 2Bb |
| chr25_-_19219807 | 0.06 |
ENSDART00000183577
|
acanb
|
aggrecan b |
| chr17_-_36513936 | 0.05 |
ENSDART00000165075
|
DCDC2C
|
si:dkey-25g12.4 |
| chr10_+_40548616 | 0.04 |
ENSDART00000147894
|
si:ch211-238p8.10
|
si:ch211-238p8.10 |
| chr23_-_36316352 | 0.04 |
ENSDART00000014840
|
nfe2
|
nuclear factor, erythroid 2 |
| chr3_+_13637383 | 0.04 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr16_+_17715243 | 0.03 |
ENSDART00000149437
ENSDART00000149596 |
si:dkey-87o1.2
|
si:dkey-87o1.2 |
| chr6_+_32415132 | 0.03 |
ENSDART00000155790
|
kank4
|
KN motif and ankyrin repeat domains 4 |
| chr8_+_42917515 | 0.03 |
ENSDART00000021715
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr7_+_48675347 | 0.03 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr21_+_19517492 | 0.03 |
ENSDART00000123168
ENSDART00000187993 |
gzmk
|
granzyme K |
| chr21_+_21812311 | 0.02 |
ENSDART00000151253
|
neu3.4
|
sialidase 3 (membrane sialidase), tandem duplicate 4 |
| chr5_+_52167986 | 0.02 |
ENSDART00000162256
ENSDART00000073626 |
slc14a2
|
solute carrier family 14 (urea transporter), member 2 |
| chr8_+_26254903 | 0.01 |
ENSDART00000113763
|
slc26a6
|
solute carrier family 26, member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rorc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.3 | 1.6 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.2 | 3.1 | GO:0090382 | phagosome maturation(GO:0090382) |
| 0.2 | 1.6 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.4 | GO:0009838 | abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.6 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.1 | 1.4 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 1.9 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.7 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 0.3 | GO:0070920 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 2.4 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.1 | 0.5 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.2 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.2 | GO:0070375 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.4 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.8 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.6 | GO:0042982 | amyloid precursor protein metabolic process(GO:0042982) |
| 0.0 | 0.3 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.6 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 1.0 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.8 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 1.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.7 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 4.8 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.3 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 1.2 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.0 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 3.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 1.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 1.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.6 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.7 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.5 | 1.4 | GO:0005131 | growth hormone receptor binding(GO:0005131) growth hormone activity(GO:0070186) |
| 0.3 | 1.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.2 | 1.0 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 1.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 2.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.2 | 0.9 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.2 | 1.6 | GO:0003834 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.4 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 1.0 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.6 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.1 | 9.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.4 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 3.6 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 1.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 5.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.1 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.8 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |